ADVERTISEMENTS:
In this article we will discuss about:- 1. Morphology of Phage Lambda 2. DNA and Gene Organization of Phage Lambda 3. Life Cycles of Phage Lambda 4. Genetic Map of Phage Lambda 5. Choice between Lytic and Lysogenic Cycles.
Morphology of Phage Lambda:
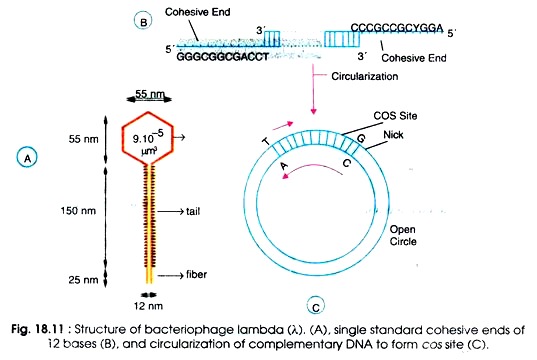
Morphological structure of phage λ is given in Fig. 18.11A. The head has 20 faces. A 20- faces 3-D picture is called an icosahedron. The head is made of protein of several types and contains a 46,500 bp long genomic (g) DNA. The phage λ contains double stranded circular DNA of about 17 µm in length packed in protein head of capsid. The head is 55 nm in diameter consisting of 300-600 capsomers (subunits) of 37,500 Daltons.
ADVERTISEMENTS:
The capsomers are arranged in clusters of 5 and 6 subunits i.e. pentamers and hexamers. The head is joined to a non-contractile 180 µm long tail by a connector. The tail consists of 35 stacked discs. It ends in a fiber.
There is a hole in capsid through which passes this narrow neck portion expanding into a knob like structure inside. The tail possesses a thin tail fibre (25 nm long) at its end which recognises the hosts. Also the tail consists of about 35 stacked discs or annuli. Unlike T-even phage, it is a simple structure devoid of the tail sheath.
Bacteriophage A falls under the family Siphoviridae of the Group I (dsDNA viruses). Phage lambda is a virus of E. coli K12 which after entering inside host cell normally does not kill it in-spite of being capable of destroying the host.
Therefore, it leads its life cycle in two different ways, one as virulent virus and the second as non-virulent. The virulent phase is called lytic cycle and the non- virulent as temperate or lysogenic one, and the respective viruses as virulent phage and temperate phage, respectively. The other temperate lamboid phages are 21, Ø80, Ø81, 424, 434, etc.
DNA and Gene Organization of Phage Lambda:
Lambda DNA is a linear and double stranded duplex of about 17 µm in length. It consists of 48, 514 base pairs of known sequence. Both the ends of 5′ terminus consists of 12 bases which extend beyond the 3′ terminus nucleotide. This results in single stranded complementary region commonly called cohesive ends.
The cohesive ends form base-pairs and can easily circularize. Consequently a circular DNA with two single strand breaks are formed. The double stranded region formed after base pairing of complementary nucleotides is designated as COS.
The 12 nucleotides of cohesive ends and process of circularization are shown in Fig. 18.11B-C. The events of circularization occurs after injection of phage DNA into E.coli cell where the bacterial enzyme, i.e., E.coli DNA ligase, converts the molecule to a covalently sealed circle.
Life Cycles of Phage Lambda:
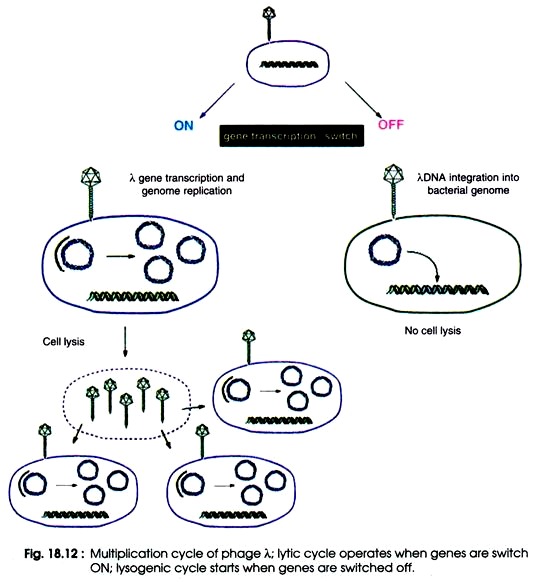
Upon adsorption on the lamb receptor of the host cell, lambda gDNA is injected through the tail which forms a hollow tube through which the DNA passes to the cell. The phage λ leads two life cycles, the lytic cycle and the lysogenic cycle after injecting its DNA into E.coli cell.
In the lytic cycle, phage genes are expressed and DNA is replicated resulting in production of several phage particles. The lytic cycle ends with lysis of E.coli cells and liberation of phage particles. This lytic cycle is a virulent or Sin-temperate where phage multiplies into several particle (Fig. 18.12).
In addition, the lysogenic cycle results in integration of phage DNA with bacterial chromosome and becomes a part of host DNA. It replicates along with bacterial chromosome and is inherited into progenies. The phage DNA integrated with bacterial chromosome is called prophage.
The prophage is non-virulent and termed as temperate phage. The bacteria containing prophage are called lysogenic bacteria, and the prophage stage of viruses as lysogenic viruses. After treatment of lysogenic bacteria with UV light. X-rays or mitomycin, the prophage can be separated from bacterial chromosomes and enter the lytic cycle. This process is known as induction (Fig. 18.12).
Genetic Map of Phage Lambda:
The genetic map of phage λ is given in Fig 18.13. The remarkable characteristics of the map is the clustering of genes according to their functions. For example the head and tail synthesis, replication and recombination genes are arranged in four distinct clusters. These genes can also be grouped into three major operons viz. right operon, left operon and immunity operon.
ADVERTISEMENTS:
The right operon is involved in the vegetative function of the phage e.g. head synthesis, tail synthesis and DNA replication leading lytic cycle. The left operon is associated with integration and recombination events of lysognic cycle.
The immunity operon products interact with DNA and decide whether the phage will initiate lytic cycle or lysogenic cycle. Singer et al (1977) have given the nucleotide sequence of ØX174. Genetic map of bacteriophage has been given by Echols and Murialdo (1978).
Function of some important genes is summarized in Table 18.4 and briefly described below.
(i) Head Synthesis Genes:
ADVERTISEMENTS:
At the left end of phage genome the head genes viz. A, W, B, C, D, E are located which are associated with phage DNA maturation and head proteins.
(ii) Tail Synthesis Genes:
The genes F, Z, U, V, G, H, M, L, K, I, J are clustered just right to head genes and code for tail proteins.
ADVERTISEMENTS:
(iii) Excision and Integration Genes:
The gene xis codes protein that excises the phage DNA from the bacterial chromosomes, and int coded protein is involved in integration of phage DNA into the bacterial chromosome.
(iv) Recombination:
The two genes int and xis codes at att P for site-specific recombination. The three red genes code for three proteins at normal frequency for general recombination. The redL codes for exonuclease, red B for beta-protein and red V for gamma protein. The gamma protein inhibits exonuclease V.
(v) Positive Regulation Gene:
The genes N and R are the positive regulation genes. The proteins code4by these genes increase the rate of transcription of other genes. Protein coded by N gene induces the transcription of cll, Q, P, A, red, gam, xis and int, whereas the protein coded by Q gene stimulates the transcription of head, tail and lysis genes. The N and Q genes are also required in plaque formation, in the absence of which the number of phage particles would be less but not zero.
(vi) Negative Regulation Genes:
The cl gene acts as a repressor and its product maintains the prophage in the lysogenic form in bacterial host. Moreover, the cll and cIII assist the d gene in lysogeny. The proteins encoded by cro binds to PL and PR and reduce the expression of cl, N, red and xis genes.
The interactions between Q proteins encoded by cro and phage repressor occur in host cell and the result decides the operation of lytic or lysogenic cycle. The choice between lysogeny and lysis has been discussed in the preceeding section.
(vii) DNA Synthesis Genes:
ADVERTISEMENTS:
The two genes O and P are involved in synthesis of phage DNA. The origin of DNA replication lies within the coding sequence for gene Q which encodes a protein for initiation of DNA replication, and the gene that generates the cohesive ends is located adjacent to one of the ends. The function of gene N is required in transcriptional process of these genes.
(viii) Lysis Genes:
The S and R genes control the lysis of bacterial cell envelope which occurs at the end of lytic cycle.
Choice between Lytic and Lysogenic Cycles:
Soon after circularization of genome and start of transcription, the gpcll and gpcIII accumulate. The gpcll binds to PRE (promoter for repressor establishment) and stimulates binding of RNA polymerase (Fig 18.14A). The gpcIII protects gpcll from degradation by host nucleases.
Lambda repressor (gpcl) is rapidly synthesized (B), binds to OL and OR, and inhibits the synthesis of mRNA and production of gpcll and gpcIII (proteins) (C). The repressor activates the promoter for repressor maintenance (PRM) which induces c/gene to be transcribed continuously at a low rate. This process goes on continuously and ensures the stable lysogeny when it is established (C).
During the course of time the gpcro also accumulates. It binds to OL and OR, turns of the transcription repressor gene cl and represses PRM function (D). The repressor (gpcl) can block cro transcription. Therefore, there is a race between the production of gpcl and gpcro proteins.
ADVERTISEMENTS:
The detail of this competition is not yet clear but the environmental factors influence the result of this race for choice of the two cycles. If the repressor wins the competitions, the circular DNA is inserted into the E. coli genome. The amount of gpcro and the outcome of competition with gpcl decide the establishment of lysogenic or lytic pathways.
Fig. 18.14 : Choice between lysis and lysogeny.