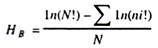ADVERTISEMENTS:
Here is a compilation of term papers on ‘Enzymes’ for class 9, 10, 11 and 12. Find paragraphs, long and short term papers on ‘Enzymes’ especially written for school and college students.
Term Paper on Enzymes
Term Paper Contents:
- Term Paper on the Meaning of Enzymes
- Term Paper on the Properties of Enzymes
- Term Paper on the Models of Enzyme Action
- Term Paper on the Types of Enzymes
- Term Paper on the Classification and Nomenclature of Enzymes
- Term Paper on Bio-Catalysis
- Term Paper on the Determination of Enzymatic Activity (Catalytic Power)
- Term Paper on the Factors that Affect Catalytic Activity of Enzymes
- Term Paper on the Inhibition of Enzyme Activity
- Term Paper on Nontraditional Enzymes
ADVERTISEMENTS:
Term Paper # 1. Meaning of Enzymes:
ADVERTISEMENTS:
Enzymes are proteins that speed up biochemical reactions without being consumed or changed by the reaction. They are found throughout nature; in our bodies, in the environment, and in all living things.
Without enzymes, life would not be possible. Enzymes speed up chemical and biochemical reactions, and this process is called catalysis. The substances upon which enzymes work when performing catalysis are known as substrates, and each enzyme will only fit and act upon a specific set of substrates.
The chemical reactions occurring within the cytoplasm of cells are also under the control of catalysts – biological catalysts called enzymes.
ADVERTISEMENTS:
Catalysts are particular chemicals that can affect how quickly chemical reactions occur. Only very small amounts of catalysts are needed to do their job. They are not used up or changed by the reactions they effect, which means they can go on working, continuously catalysing the same reaction, so long as the reactant molecules (the substrate molecules) are present. Catalysts usually speed up chemical reactions.
There are so many enzymes that it would be impossible to name them all. In fact, scientists have yet to discover many enzymes, or fully understand their structure and properties. On the other hand, many other enzymes have been successfully studied and applied to industrial and commercial uses.
Term Paper #
2. Properties of Enzymes:
Special properties of enzymes are:
(i) They are all protein molecules.
(ii) They are made in the cytoplasm under instruction from genes on the chromosomes in the nucleus.
(iii) Each enzyme works at its fastest rate at one particular temperature. This is known as its optimum temperature. For most enzymes in a mammal’s body, the optimum temperature is around 37°C. As the temperature rises towards 37°C, the rate of enzyme activity speeds up. If the temperature rises above the optimum, the rate of activity begins to slow down. At around 60°C, the enzyme is destroyed or ‘denatured’ (not ‘killed’ – it was never alive). The rate of the reaction falls to (almost) 0.
(iv) Each enzyme works at its fastest rate at one particular pH. This is known as its optimum pH. If the pH level falls either side of the optimum, the rate of enzyme activity gradually decreases. For most enzymes in a mammal’s body, the optimum is pH 7, or slightly above. However, the optimum for each enzyme depends on where it is found in the body. For example, the enzyme protease found in the stomach, where it is highly acidic, is most effective at the low pH 1.5.
Remember:
ADVERTISEMENTS:
pH is a measure of the degree of acidity or alkalinity. A pH of 7 is neutral, below 7 is acidic and above 7 is alkaline.

Term Paper #
3. Models of Enzyme Action:
Two models have been proposed to explain the modes of enzyme action—lock and key model and induced-fit model.
1. Lock and Key Model:
ADVERTISEMENTS:
Lock and key model and respective hypothesis was given by the Emil Fischer in 1874. This model explains that both enzyme and substrate molecules have matching shapes like ‘lock’ and ‘key’ in the region of active sites. It is because the enzyme and the substrate interact by means of short-range forces that require close contact. Just as a lock can be opened by its specific key, a substrate molecule can be acted upon by a particular enzyme (Fig. 27.3).
The job of an enzyme (e.g. those used in digestion) is often to break down a large molecule (the substrate molecule) into smaller molecules (the product). The ‘lock and key’ hypothesis suggests how this may be achieved.
Each enzyme is a molecule with a specific shape. On part of its surface is the active site (the ‘lock’) – a section where its substrate molecule (the ‘key’) fits exactly. When the substrate molecule is in position in the active site, the enzyme slightly stresses (or ‘bends’) the substrate, splitting it into two product molecules. The product molecules drift away from the enzyme molecule, leaving its active site free to operate again.
ADVERTISEMENTS:
By including a molecule of water in this process, the newly exposed ends of the product molecules are ‘sealed’ so they will not re-join. This type of enzyme- controlled reaction, common in digestion, is known as hydrolysis.
The ‘lock and key’ hypothesis explains enzyme action because:
(i) Only the correct enzyme-substrate combination can work.
ADVERTISEMENTS:
(ii) Higher temperatures make the enzyme and substrate molecules move more quickly. This also means the substrate molecules enter the active site of the enzyme, and the product molecules leave the active site of the enzyme, more quickly.
(iii) Extreme heat causes the atoms of the enzyme molecule to move about so violently that they change position relative to one another. This changes the shape of the active site, and the enzyme stops doing its job.
(iv) Changes in pH may alter the shape of large molecules like proteins. If that protein is an enzyme, the shape of the active site may be changed, and the enzyme will work less efficiently.
2. Induced-Fit Model:
Induced-fit model (Fig. 27.4) was proposed by Daniel E. Koshland in 1958 by modifying lock and key model. An important but unfortunate feature of Fischer’s lock and key model is the rigidity of the active site; the active site is presumed to be pre-shaped to fit the substrate. Koshland, however, presumed that the enzyme molecule does not retain its original shape and structure.
On the other hand, the contact of the substrate induces some configurationally or geometrical changes in the active site of the enzyme molecule. We now know that the enzymes are flexible and that the shapes of the active sites can be markedly modified by the binding of substrate.
Term Paper #
4. Types of Enzymes:
A few basic enzyme types are briefly described as follows:
In the absence of enzymes, chemical reactions occur only when molecules collide while in proper alignment with each other. Because molecules are bumping into each other randomly, chemical reactions are essentially due to chance events.
This sometimes results in reactions that occur very slowly, or reactions that do not occur at all. Enzymes act like tiny molecular machines to ensure that molecules come into contact with each other and react.
Like a key fitting into a lock, chemical molecules fit into pocket-like structures located on an enzyme. These pockets hold the molecules in a position that will allow them to react with each other, ensuring that they are close enough together and aligned properly for a reaction to occur.
ADVERTISEMENTS:
In this way, enzymes speed up reactions. The enzymes are not changed themselves by the reaction. When the reaction is complete, enzymes release the product(s) and are ready to bring together more molecules and catalyse more reactions.
Biotechnology can provide an unlimited and pure source of enzymes as an alternative to the harsh chemicals traditionally used in industry for accelerating chemical reactions. Enzymes are found in naturally occurring microorganisms, such as bacteria, fungi, and yeast, all of which may or may not be genetically modified.
Large quantities of enzymes are often needed for industrial use, so these microorganisms are multiplied through a process called fermentation. When enzymes or the microorganisms used to create them are no longer needed, they are destroyed through exposure to heat or safe organic/inorganic materials.
Term Paper #
5. Classification and Nomenclature of Enzymes:
Traditionally, enzymes often were named by adding the suffixase to the name of the substrate upon which they acted, e.g., the name phosphatase for enzymes hydrolyzing phosphoryl groups from organic phosphate compounds and the name urease for enzymes hydrolyzing urea.
Certain enzymes were so named that they little resembled to their activity, e.g., the name catalase for enzymes decomposing peroxide and the name proteases for proteolytic activity.
To avoid confusions arising from these trivial designations, however, the International Union of Biochemistry established an International Commission of Enzymes in 1964 to create a systematic basis for enzyme classification and nomenclature.
Although the trivial names for many enzymes remain in use, the Enzyme Commission classified and named enzymes according to the reaction they catalyse:
(i) Enzyme Commission Recommendation:
Enzyme Commission divided the reactions into six major classes numbered 1 through 6. Within each class are subclasses, and under each subclass are sub-subclasses within which individual enzymes are listed. Classes, subclasses, sub-subclasses, and individual entries are each numbered, so that a series of four numbers serves to specify a particular enzyme.
The series of four numbers is preceded by the letters EC for Enzyme Commission. For example, EC 2.7.1.1 is used to designate the formal systematic name of the enzyme ATP: glucose phosphotransferase, which catalyses the reaction:
ATP + D-glucose → ADP + D-glucose 6-phosphate
The reaction indicates that the enzyme ATP: glucose phosphotransferase catalyses the transfer of a phosphoryl group from ATP to glucose. The first digit (2) of its Enzyme Commission number (EC 2.7.1.1) denotes the class name (transferase); the second digit (7) denotes the subclass (phosphotransferase); the third digit (1) denotes the sub-subclass (hydroxy I group as acceptor); and the fourth digit (1) denotes D-glucose as the phosphoryl-group acceptor.
These designations are very cumbersome, so in everyday usage, trivial names are employed frequently. Kinase is a trivial term for enzymes that are ATP-dependent phosphortransferases. The glucose-specific enzyme (EC 2.7.1.2) is called glucokinase and the nonspecific (EC 2.7.1.1) is known as hexokinase. However, the classification number of an enzyme is preferably used when the precise identity of the enzyme might be ambiguous.
(ii) Six Major Classes of Enzymes:
The six major classes of enzymes as recommended by the Enzyme Commission are the following:
Class 1:
Oxidoreductases are involved in redox reactions, i.e., transfer of hydrogen or oxygen atoms between molecules. This class includes dehydrogenases (hydride transfer), oxidases (e– transfer to O2), oxigenases (oxygen atom transfer from O2), and peroxidases (e– transfer to peroxides).
Example:
Glucose oxidase (EC 1.1.3.4).
Class 2:
Transferases catalyse the transfer of an atom or group of atoms (like acyl-, alkyl- and glycosyl- groups) between two molecules. The transferred groups are different from those transferred by the other classes of enzymes like oxidoreductases, etc.
Example:
Aspartate aminotransferase (EC 2.6.1.1).
Class 3:
Hydrolases are those enzymes, which catalyze hydrolytic reactions (and their reversals); this class includes esterases, glycosidases, proteases and lipases.
Example:
Chymosin or rennin (EC 3.4.23.4).
Class 4:
Lyases are involved in elimination reactions resulting in the removal of a group of atoms from the substrate molecule. This class includes aldolases, decarboxylases, dehydratases and some pectinases.
Example:
Histidine ammonia lyase (EC 4.3.1.3).
Class 5:
Isomerases catalyse the formation of isomers of molecules; they include epimerases, racemases and intramolecular transferases.
Example:
Xylope isomerase (EC 5.3.1.5).
Class 6:
Ligases, Synthases or Synthetases catalyze the formation of covalcnt bonds between two molecules utilizing the energy obtained from hydrolysis of a nucleoside triphosphate like ATP or GTP.
Example:
Glutathione synthase (EC 6.3.2.3).
Term Paper #
6. Bio-Catalysis:
Biocatalysis, the enzymatic catalysis of chemical reactions, is essential to living organisms. Enzymes accelerate chemical reactions by factors of as much as a million or more. In fact, most chemical reactions in biological systems do not occur at perceptible rates in the absence of enzymes. Even a reaction as simple as the hydration of CO2 is catalyzed by an enzyme, namely, carbonic anhydrase.
Following is a concise account in reference to how enzymes work as biocatalysts in living systems:
Enzymes Lower the Activation Energy of the Reaction:
Enzymes increase the rate of a reaction without affecting its equilibrium. This they do by lowering the activation energy of the reaction. Let us first understand the concept of activation energy, and then the mechanism lowering the activation energy by enzymes.
1. Activation Energy:
Almost all biochemical reactions in a cell do not start automatically. It is because the substrate (reactant) molecules have an energy barrier and remain “nonreactive”. This state is called ground state.
The substrate molecules remain in ground state on account of:
(i) Mutual repulsion due to pre-sense of electrons on their surfaces,
(ii) Solvation or holding of reactants in solution form by hydrogen bonds, and
(iii) Reaction sites of the reaction molecules being small, precise collisions do not take place.
To undergo reaction, the molecules must overcome the ground state (i.e., the “non-reactive” state); therefore, they must be raised to a higher energy level to become “reactive”. The reactive state of substrate is called transition state.
The transition state is simply a fleeting molecular moment in which events such as bond breakage, bond formation, and charge development take place. The difference between the energy levels of the ground state (nonreactive state) and the transition state (reactive state) is called the activation energy (Fig. 27.2).
2. Lowering the Activation Energy by Enzymes:
A reaction taking place without a catalyst requires a high amount of activation energy. When the same reaction is catalyzed by an enzyme, the required amount of activation energy is remarkably lowered. For example, decomposition of H2O2 without a catalyst requires 18,000 cal/mol energy; this is lowered to 2,000 cal/mol when the enzyme catalase catalyzes the reaction.
The rate at which the substrate (reactant) A is converted to the product B is dependent not on the concentration of molecules of substrate A but on the number of molecules of substrate A with an energy greater than the energy barrier. The molecules of substrate A in solution have different energies since they are continuously moving about hitting one another.
When a molecule through collision or otherwise obtains its required activation energy and crosses the energy barrier, it becomes reactive and reacts to be converted to product B. If this reaction is catalyzed by an enzyme, the required activation energy is lowered.
The lowering of activation energy by the enzyme is due to a combination of the enzyme with the substrate. The combination of enzyme and substrate (i.e., enzyme-substrate complex) creates a new reaction pathway whose transition state energy is lower than that of the reaction in the absence of enzyme (Fig. 27.2).
The lowering of the transition state energy in the new reduction pathway can be attributed to the following:
(i) Some weak interactions are formed in the enzyme-substrate-complex. The free energy (binding energy) released by the formation of these interactions compensates much of the activation energy difference between the un-catalyzed and the enzyme-catalyzed reactions.
(ii) Functional groups of the enzyme’s amino acid residues may interact with the various bonds in the substrate, thereby weakening them to allow their conversion into product.
(iii) Enzymes have ability to cause distortion in the substrate molecules thereby straining and weakening specific bonds, leading to faster product formation.
(iv) In two substrates involving reactions such as, A + B (substrates) Energy → C + D (products), the enzyme’s active site provides bindings sites for both substrates. This allows them to interact more closely and appropriately.
(v) A number of catalytic mechanisms are possible due to the wide array of functional groups present in the R-groups of amino acid residues of enzymes. These add to the catalytic power of the enzyme.
The lowering of activation energy means that more substrate molecules have the required energy to reach the transition state and therefore the rate of reaction is increased.
Term Paper # 7. Determination of Enzymatic Activity (Catalytic Power):
Enzyme Kinetics:
When the enzyme is first mixed with a large excess of substrate, there is an initial period, the pre-steady state, during which the concentration of ES-complex (enzyme-substrate-complex) builds up. This period proceeds very quickly and is usually too short to be easily observed.
The velocity of the reaction quickly achieves a steady-state in which ES-complex remains approximately constant over time and since it has reached a maximum value point, it is referred to as maximum velocity (Vmax). The substrate concentration at maximum velocity is very high and saturates all the active sites of the enzyme allowing it to turn out products at the maximum rate.
Enzyme-catalyzed reactions must be performed at appropriate pH (using a buffer) and optimum temperature (usually between 20- 37°C). It is necessary because the enzymes are active as catalysts only if the conformation (3-D structure) remains intact. If the value of pH or temperature is altered, the enzyme undergoes denaturation, and its 3-D structure is destroyed.
The enzyme-activity (the catalytic power of enzyme) can be determined experimentally by allowing an enzyme to interact with an excess of its substrate for a given period of time and then determine the amount of product formed by some chemical or physical property such as colour, adsorption spectrometry, radioactivity, etc.
Now to determine the initial velocity (V0) of an enzyme-catalyzed reaction at different concentrations of its substrate, a graph is plotted (Fig. 27.5) of V0 versus [S], where V0 is the initial velocity of an enzyme catalyzed reaction and [S] is the molar concentration of the substrate.
The curve expressing the relationship between V0 and [S] has the same general shape for most enzymes (it approaches a rectangular hyperbola), which can be expressed mathematically by the Michaelis-Menten equation given by Leonar Michaelis and Maud Menten in 1913.
Michaelis and Menten derived this equation starting from their basic hypothesis that the rate limiting step in enzyme-catalysed reactions is the breakdown of the ES-complex to product and free energy.
This equation is as follows:
The important terms of this equation are [S], V0, Vmax, and a constant called Michaelis constant, Km. [S], V0, and Vmax are mentioned earlier, Km represents the substrate concentration at which the velocity is half the value of Vmax. The value of Km is inversely proportional to the affinity of the enzyme for its substrate. A low value of Km means the enzyme has a high affinity for its substrate.
From the Vmax value, one can calculate a value called ‘turnover number’ because Vmax is equal to the turnover number and the total concentration of enzyme.
Turnover number explains how many molecules of substrate are converted to product per second. Turnover numbers for most enzymes are between 1 and 104 per second. Larger the turnover number, better the enzyme’s catalytic power. Table 27.2 shows the maximum turnover numbers of some enzymes.
Term Paper # 8
. Factors that Affect Catalytic Activity of Enzymes:
Enzymes activity varies greatly with changes in environmental factors such as substrate concentration, and alterations in pH and temperature.
1. Substrate Concentration:
The concentration of a substrate in the medium affects the activity of the enzyme that catalyses it. At very low concentrations, an enzyme makes product slowly because it seldom contacts a substrate molecule.
If more substrate molecules are present, an enzyme binds substrate more often, and the velocity of the reaction, which is usually expressed in terms of the rate of product formation, is greater than at a lower substrate concentration.
Thus, the rate of an enzyme-catalysed reaction increases with increase in the substrate concentration but, this increase ceases after a limit. Any further increases in substrate concentration do not result in a greater reaction velocity because the available enzyme molecules are binding substrate and converting it to product as rapidly as possible, i.e., the enzyme is saturated with substrate and operating at maximal velocity (Vmax).
When we plot a graph taking velocity of enzyme-catalysed reaction and substrate concentration, we can find that the resulting substrate concentration curve is a hyperbola (Fig. 27.6).
2. Alterations in pH:
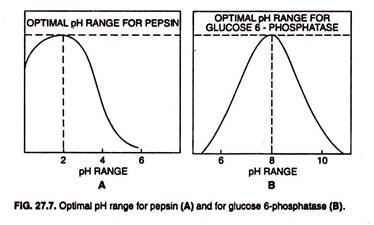
Each enzyme has an optimum pH (or pH range) and functions most rapidly at that. The optimum pH range of the most enzymes is 4-9; for pepsin it is 2 and for glucose 6-phophatase it is 8. When the pH deviates too greatly from an enzyme’s optimum, the activity of enzyme slows (Fig. 27.7) and the enzyme may be damaged.
This so happens because the amino acid side chains in the active site of an enzyme may act as weak acids and bases with critical functions that depend on their maintaining a certain state of ionisation, and elsewhere in the enzyme, ionised side chains may play an essential role in the interactions that maintain enzyme structure.
Removing a proton from a His residue of an amino acid side chain, for instance, might eliminate an ionic interaction essential for stabilisation of the active conformation of the enzyme.
The pH range over which an enzyme undergoes changes in activity can provide a clue to what amino acid is involved in the reaction. A change in activity near pH 7.0, for example, often reflects the removal of a proton from His residue of histidine amino acid.
3. Alterations in Temperature:
Like pH, enzymes have temperature optima for maximum activity. Below the temperature optima, every increase in temperature by 10°C normally doubles the rate of an enzyme-catalyzed chemical reaction but, if the temperature goes too much above the optimum, an enzyme’s structure is disrupted and its activity lost (Fig. 27.8).
This phenomenon is called denaturation. The temperature optima of a microorganism’s enzymes often reflect the temperature of its habitat. Although great majority of microorganisms loss the activity of their enzymes at 40°C- 60°C due to denaturation, many a number of bacteria and archaebacteria, not surprisingly, grow best at extremely high temperatures as their enzymes have very high temperature optima and great heat stability.
Term Paper # 9
. Inhibition of Enzyme Activity:
The activity of enzymes can be reduced or stopped by molecular agents called inhibitors; the inhibitors may be specific small molecules and ions. Inhibition of enzyme action serves as a major control mechanism in biological systems and, therefore, the study of enzyme inhibitors provides valuable information about enzyme mechanisms and helps define important metabolic pathways in biological systems.
Inhibition of enzyme action can be studied under the heads:
(1) Reversible inhibition and
(2) Irriversible inhibition.
(1) Reversible Inhibition:
Reversible inhibition is of temporary nature, and is characterized by a rapid dissociation of the enzyme- inhibitor complex. It can be overcome by withdrawal of the inhibitor due to blocking of active site or binding of linkages required for maintenance of active site. Dilution and dialysis reduces or eliminates the effect of reversible inhibition. Reversible inhibition may be competitive, uncompetitive, or noncompetitive.
a. Competitive Inhibition:
Competitive inhibition is the common type of reversible inhibition. In it the inhibitor resembles the substrate and competes with the latter for the active site of an enzyme. When the inhibitor occupies the active site of the enzyme, it prevents binding of the substrate to the enzyme (Fig. 27.9).
Competitive inhibitors are often compounds that resemble the substrate and combine with the enzyme to form an enzyme-inhibitor-complex (ET-complex), but without leading to catalysis.
For example, methotrexate is a structural analog of tetrahydrofolate, a coenzyme for the enzyme dihydrofolate reductase, which plays a role in the biosynthesis of purines and pyrimidines. The methotrexate binds to dihydrofolate reductase 1000-fold more tightly than does the natural substrate and inhibits nucleotide base synthesis.
Since the competitive inhibition is reversible in nature, the competition between the substrate and inhibitor to bind enzyme can be overcome by increasing the substrate concentration, when substrate(s) concentration far exceeds inhibitor (I) concentration, the probability that an inhibitor molecule will bind to the enzyme is minimized, and the reaction exhibits a normal maximum initial velocity (Vmax).
Competitive inhibitors are very useful in certain medical therapies to treat patients.
Following are the two examples.
(i) Sulfa drugs like sulfanilamide are analog to p-amino-benzoate, a molecule used in the synthesis of the coenzyme folic acid. The sulfa drugs, when enter a bacterial cell; compete with p-aminobenzoate for the catalytic site of an enzyme involved in folic acid synthesis.
This blocks the production of folic acid and inhibits bacterial growth or causes death of the pathogenic bacteria. In contrast, humans do not synthesise folic acid and must obtain it in the diet; therefore, sulfa drugs do not affect them.
(ii) A patient who has ingested methanol can be treated using competitive inhibitor. Methanol is converted to formaldehyde by liver enzyme alcohol dehydrogenase-, the formaldehyde damages many tissues and may cause blindness. In this case the ethanol can be used as competitive inhibitor.
The ethanol competes effectively with methanol as an alternative substrate for alcohol dehydrogenase and is converted to acetaldehyde by the enzyme. This prevents methanol conversion to formaldehyde thus lessening the danger. However, in the meantime, the kidneys filter out the methanol to be excreted harmlessly in the urine.
b. Uncompetitive Inhibition:
Uncompetitive inhibition is the reversible inhibition in which the inhibitor binds at a site distinct from the active site of enzyme for the substrate and, unlike a competitive inhibitor, binds only to the enzyme- substrate complex (ES complex) and not to free enzyme (Fig. 27.10).
An uncompetitive inhibitor acts by decreasing the turnover number rather than by diminishing the proportion of enzyme molecules that are bound to substrate. Uncompetitive inhibition, in contrast with competitive inhibition, cannot be overcome by increasing the substrate concentration because the substrate is more effectively bound to the enzyme in the presence of the inhibitor.
c. Noncompetitive Inhibition:
Noncompetitive inhibition is such a reversible inhibition in which the inhibitor binds to an enzyme whether or not its active site is occupied by the substrate, i.e., the inhibitor binds with either the enzyme or the enzyme- substrate complex (Fig. 27.11) Noncompetitive inhibition therefore differs from competitive inhibition in that the inhibitor can combine with enzyme-substrate complex, and substrate can combine with enzyme- inhibitor complex to form in both instances the enzyme-inhibitor-substrate complex.
Since the inhibitor-binding site is not identical to the active site, nor does it modify the active site directly, the noncompetitive inhibitor decreases the maximum velocity (Vmax) of an enzyme reaction without affecting the Km (Michaelis constant). Noncompetitive inhibition therefore cannot be overcome by increasing the substrate concentration.
(2) Irreversible Inhibition:
Irreversible inhibition is of permanent nature. In it the irreversible inhibitor combines with or destroys a functional group (usually an amino acid side chain) on an enzyme that is essential for the activity of the enzyme, or that forms a particularly stable non-covalent association. Suicide inactivators (also called mechanism-based inactivators) are irreversible inhibitors. They remain unreactive until they bind to the active site of a specific enzyme.
In reactive state, a suicide inactivator carries out the first few chemical steps of the normal enzyme reaction, but instead of being transformed into the normal product, the indicator is converted to a very reactive compound that combines irreversibly with the enzyme. Some important drugs are also irriversible inhibitors.
Penicillin binds covalently to enzyme trans-peptidase, modifies it, and thereby prevents the synthesis of bacterial cell walls. Aspirin acts by covalently modifying the enzyme cyclooxygenase, thereby reducing the synthesis of inflammatory signals. Heavy metals (e.g., Ag+, Hg2+, As2+) and iodoacetic acid combine with -SH group and destroy enzyme structure.
Term Paper #
10. Nontraditional Enzymes:
A great majority of the enzymes are highly evolved and extremely specific proteins.
ADVERTISEMENTS:
However, two other forms of biological catalysts are now known; these are:
(i) Abzymes and
(ii) Ribozymes.
(i) Abzymes:
Abzymes are antibodies that catalyze specific chemical reactions, i.e., function as enzymes. Antibodies, by definition, have evolved to recognize and bind to the ground states of the molecules they are specific to. In contrast, enzymes have binding sites that preferentially bind to the transition states of their substrate molecules. For this reason, enzymes are not produced naturally.
A catalytic antibody is produced in response to molecules that have a structure similar to the proposed/expected transition state of the substrate for the reaction to catalyse which the antibody (abzyme) is sought. Such molecules are, in fact, stable analogues of the transition states of the concerned substrates.
The fact that such catalytic antibodies are in fact produced is the strongest evidence in support of the transition theory of enzyme action. Abzymes have been produced that catalyze such reactions as hydrolysis, e.g., of esters and carbonates, cleavage of amide bond, and bio-molecular chemical reactions. Some abzymes incorporate metal ion cofactors like conventional protein enzymes.
(ii) Ribozymes:
RNA molecules that have the capability of catalyzing chemical reactions are called ribozymes. Ribozymes are true catalysts in that they enhance the rate of chemical reactions without any net change to themselves. They are capable of turnover, i.e., recycling, and show kinetics typical of enzymes. Ribozymes are so far known to catalyze reactions like cleavage of RNA, cleavage of DNA, etc.
Ribozymes are able to cut and splice themselves into a form that can catalyze the cleavage of other RNA/DNA molecules. The catalytic ability of ribozymes arises due to their three dimensional structures, which is able to generate in them the substrate- specific binding site.
This is similar to enzymes in which the substrate specific binding site is produced by their three-dimensional structure. It may be emphasized that all biological catalysts must possess the ability to bind to their specific substrates.
Ribozymes offer exciting possibilities of varied applications, e.g., in human, animal and plant disease control. It has been suggested that RNA was the first macromolecule on earth, which catalyzed its own replication, and it subsequently developed other catalytic capabilities.
Much later, RNA acquired the ability to catalyze protein synthesis. In due course of time, proteins became the primary biological catalysts since they were capable of catalyzing a much larger variety of reactions more efficiently than ribozymes.
The first DNA molecules are thought to have originated by reverse transcription of RNA molecules. Ultimately, DNA becomes the primary genetic material in view of its greater stability than RNA.
Thus, according to this hypothesis, RNA has been dethroned from its princely status by the same two macromolecules. i.e., proteins and DNA, the synthesis of which is either catalyzed (proteins) or provided the primary information for (DNA by reverse transcription); RNA is now generally relegated to a mere secondary (although essential) role as the link between DNA and proteins.