ADVERTISEMENTS:
Let us make an in-depth study of transcription (synthesis of RNA) in prokaryotes and eukaryotes.
Transcription in Prokaryotes:
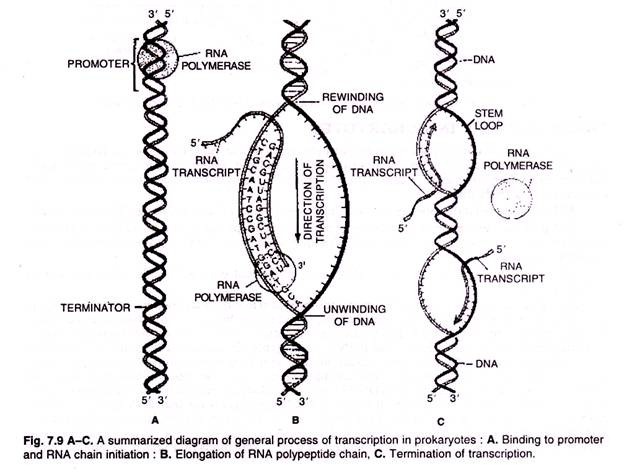
In prokaryotic organisms transcription occurs in three phases known as initiation, elongation and termination.
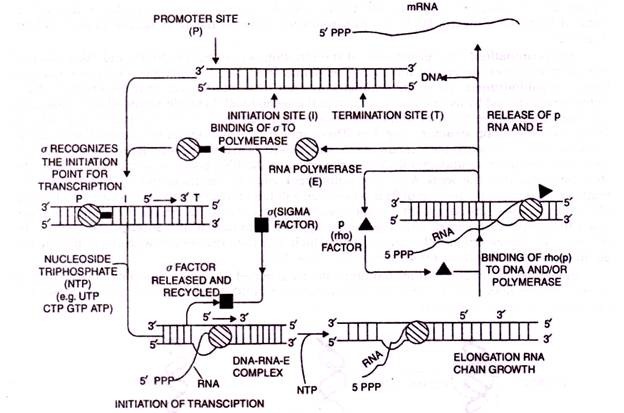
RNA is synthesized by a single RNA polymerase enzyme which contains multiple polypeptide subunits. In E. coli, the RNA polymerase has five subunits: two α, one β, one β’ and one σ subunit (α2ββ’σ). This form is called the holoenzyme. The σ subunit may dissociate from the other subunits to leave a form known as the core enzyme.
ADVERTISEMENTS:
These two forms of the RNA polymerase have different roles in transcription:
(i) Initiation:
Transcription cannot start randomly but must begin specifically at the start of a gene. Signals for the initiation of transcription occur in the promoter sequence which lies directly upstream of the transcribed sequence of the gene. The promoter contains specific DNA sequences that act as points of attachment for the RNA polymerase.
In E. coli, two sequence elements recognized by the RNA polymerase known as the -10 sequence and the -35 sequence arc present. The exact sequences can vary between promoters but all conform to an overall pattern known as the consensus sequence. The σ subunit of the RNA polymerase is responsible for recognizing and binding the promoter, probably at the -35 Box.
In the absence of the σ subunit the enzyme can still bind to DNA but binding is more random. When the enzyme binds to the promoter it initially forms a closed promoter complex in which the promoter DNA remains as a double helix. T he enzyme covers about 60 base pairs of the promoter including the -10 and -35 boxes. To allow transcription to begin, the double helix partially dissociates at the – 10 box, which is rich in weak A-1 bonds to give an open promoter complex.
ADVERTISEMENTS:
The σ subunit then dissociates from the open promoter complex leaving the core enzyme. At the same time the first two ribonucleotides bind to the DNA, the first phosphodiester bond is formed and transcription is initiated (Fig. 7.7).
(ii) Elongation:
During elongation the RNA polymerase moves along the DNA molecule melting and unwinding the double helix as it progresses. The enzyme adds ribonucleotides to the 3′ end of the growing RNA molecule with the order of addition determined by the order of the bases on the template strand.
In most cases, a leader sequence of variable length is transcribed before the coding sequence of the gene is reached. Similarly, at the end of the coding sequence a noncoding trailer sequence is transcribed before transcription ends.
During transcription only a small portion of the double helix is unwound at any one time. The unwound area contains the newly synthesized RNA base-paired with the template DNA strand and extends over 12-17 bases.
The unwound area needs to remain small because unwinding in one region necessitates over-winding in adjacent regions and this imposes strain on the DNA molecule. To overcome this problem, the RNA is released from the template DNA as it is synthesized allowing the DNA double helix to reform (Fig. 7.8).
(iii) Termination:
The termination of transcription occurs non-randomly and takes place at specific points after the end of the coding sequence. In E. coli, termination occurs at sequences known as palindromes. These are symmetrical about their middle such that the first half of the sequence is followed by its exact complement in the second half.
In single-stranded RNA molecules this feature allows the first half of the sequence to base pair with the second half to form what is known as a stem-loop structure (Fig. 7.9). These appear to act as signals for termination. In some cases the stem-loop sequence is followed by a run of 5-10 As in the DNA which form weak A-U base pairs with the newly synthesized RNA.
It is thought that the RNA polymerase pauses just after the stem-loop and that the weak A-U base pairs break causing the transcript to detach from the template. In other cases the run of As is absent and a different mechanism occurs based on binding of a protein called Rho (ρ) which disrupts base-pairing between the template and the transcript when the polymerase pauses after the stem-loop. The termination of transcription involves the release of the transcript and the core enzyme which may then re-associate with the σ subunit and go on to another round of transcription (Fig. 7.9 & 7.10).
ADVERTISEMENTS:
In many bacteria, genes of related functions are grouped together in operons. An operon acts as a single transcription unit and thus produces polycistronic mRNA. In eukaryotes, only monocistronic mRNAs are generally produced.
Fig 7.10. Diagrammatic presentation of the synthesis of RNA by E.coli polymerase
Transcription in Eukaryotes:
Transcription occurs in eukaryotes in a way similar to prokaryotes. However, initiation is more complex, termination does not involve stem-loop structures and transcription is carried out by three enzymes (RNA polymerases I, II and III) each of which transcribes a specific set of genes and functions in a slightly different way.
ADVERTISEMENTS:
RNA polymerase I transcribes genes encoding three of the four ribosomal RNAs (18S, 28S and 5.8S). RNA polymerase II enzyme transcribes genes that encode proteins. Binding of RNA polymerase II to its promoter involves several different DNA sequence elements and a number of proteins called transcription factors. RNA polymerase III transcribes a set of short genes that encode transfer RNAs and the 5S ribosomal RNA.
Unlike the situation in prokaryotic genes, transcription in eukaryotes occurs within the nucleus and mRNA moves out of the nucleus into the cytoplasm for translation. The initiation and regulation of transcription is more extensive than prokaryotes. Another major difference between prokaryotes and eukaryotes lies in the fact that the mRNA in eukaryotes is processed from the primary RNA transcript, a process called maturation.
Initially at the 5′ end a cap (consisting of 7-methyl guanosine or 7 mG) and a tail of poly A at the 3′ end are added (Fig. 7.11) The cap is a chemically modified molecule of guanosine triphosphate (GTP). The primary eukaryotic mRNA transcript is much longer and localised into the nucleus, when it is also called heterogenous nuclear RNA (hnRNA) or pre- mRNA.
ADVERTISEMENTS:
The eukaryotic primary mRNAs are made up of two types of segments; non-coding introns and the coding exons. The introns are removed by a process called RNA splicing. Of a pair of small nuclear ribonucleoprotein (SnRNPs pronounced “snurps”), one binds to 5′ splice site and the other to 3′ splice site.
A spliceosome forms because of interaction between SnRNPs and other proteins. This spliceosome uses energy of ATP to cut the RNA, releases the introns and joins two adjacent exons to produce mature mRNA. Besides, these two post-transcriptional modifications, RNA editing may also take place before translation begins.
Post-Transcription Processing:
Primary transcript is often larger than the functional RNAs. It is called heterogeneous or hnRNA especially in case of mRNA.
ADVERTISEMENTS:
Post-transcription processing is required to convert primary transcript into functional RNAs.
It is of four types:
(i) Cleavage:
Larger RNA precursors are cleaved to form smaller RNAs. Primary transcript of rRNA is 45S in eukaryotes.
It is cleaved to form the following:
Primary transcript is cleaved by ribonuclease-P (an RNA enzyme) to form 5-7 tRNA precursors,
ADVERTISEMENTS:
(ii) Splicing:
Eukaryotic transcripts possess extra segments (introns or intervening sequences). They are removed by nucleases. Ribozyme (an-RNA enzyme) is a self splicing intron involved in some of these reactions as well as catalysing polymerisation.
(iii) Terminal Additions:
Additional nucleotides are added to the ends of RNAs for specific functions, e.g., CCA segment in tRNA, cap nucleotides at 5′ end of mRNA or poly-A segments at 3′ end of mRNA.
(iv) Nucleotide Modifications:
ADVERTISEMENTS:
They are most common in tRNA-methylation (e.g., methyl cytosine, methyl guanosine), deamination (e.g., inosine from adenine), dihydrouracil, pseudouracil, etc.