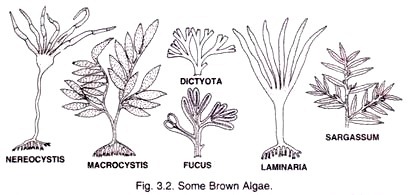
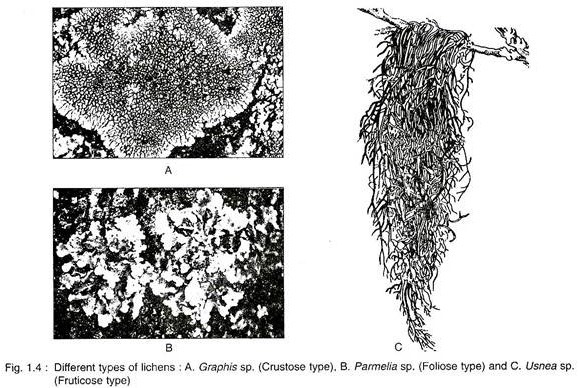
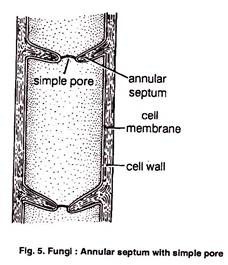
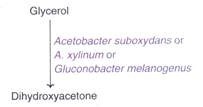
ADVERTISEMENTS:
Let us make an in-depth study of the synthesis of RNA:- Learn about- 1. Process of RNA Synthesis 2. Transcription in Prokaryotes and 3. Transcription in Eukaryotes.
Genes are expressed by transfer of genetic information from DNA to RNA. From RNA the information is used for synthesizing proteins. RNA is synthesized from a portion of one strand of DNA, which acts as a template. This process is called transcription. All RNA molecules are derived from the information permanently stored in DNA except RNA genome of certain viruses.
The base sequence of RNA strand is complementary and anti-parallel to the DNA template strand. Out of the two strands of DNA only a section of one strand acts as a template and directs the synthesis of RNA. The base sequence of other strand is the same as that of RNA. The sequence of bases of RNA leads to the corresponding sequence of amino acids of protein. Thus RNA plays a central, pivotal role in the activities of the cell.
Replication and Transcription:
ADVERTISEMENTS:
Basic process of chain elongation is similar in both replication and transcription. Nucleotides are added one by one in 5′ 3′ direction in both the cases. But there are some important differences.
During replication, the entire chromosome (both DNA strands) is copied but the transcription is more selective. One particular gene or group of genes is transcribed at any one time, producing one to numerous number of copies. Some parts of DNA are never transcribed. In transcription, RNA strand is made up of ribonucleotides rather than deoxyribonucleotides. Transcription does not require a primer but, replication needs primer for copying both strands.
Process of RNA Synthesis:
The basic features of RNA synthesis are as follows:
ADVERTISEMENTS:
1. The building blocks of RNA are four ribonucleotide 5-triphosphates. They are ATP, GTP, CTP and UTP.
In polymerization, ribonucleotides are added one-by-one. The 3′-OH group of one nucleotide reacts with 5′-triphosphate of next nucleotide, the pyrophosphate (PP) is released. This is similar to DNA polymerization in DNA replication.
2. The sequence of bases in RNA is determined by the base sequence of DNA template strand. The base sequence of RNA strand is complementary to the template strand of DNA. Thymine is replaced by Uracil in RNA.
3. Out of two strands of DNA, often one strand acts as a template for RNA synthesis. Transcription selectively copies only certain parts of genome and makes one to numerous copies of RNA. The RNA formed is antiparallel to the DNA template.
4. Unlike DNA replication, which needs RNA primer, RNA polymerase enzyme, which synthesizes RNA strand does not need a primer and can initiate transcription de novo.
RNA synthesis consists of four stages:
1. Binding of RNA polymerase enzyme to the template at a promoter site on DNA.
ADVERTISEMENTS:
2. Initiation of new strand.
3. Elongation.
4. Termination and release.
Transcription in Prokaryotes:
Length of mRNA chain depends upon the length of polypeptide chain it codes for. The mRNA in prokaryotes is generally polycistronic which codes for several proteins which represent proteins of a single metabolic pathway.
ADVERTISEMENTS:
RNA Polymerase Enzymes:
ADVERTISEMENTS:
DNA dependent RNA polymerase enzyme requires a DNA template. In addition, it requires four ribonucleotide-triphosphates which are ATP, GTP, UTP and CTP. Each nucleotide in the newly formed RNA strand is selected on the basis of Watson-Crick base pairing rule.
In prokaryotes all types of RNAs are transcribed by the same RNA polymerase enzyme RNA polymerase is one of the largest enzymes having a molecular weight 500000 daltons. When it binds DNA, it covers many bases of DNA simultaneously.
RNA polymerase consists of subunits-2α subunits, β, β and c (sigma factor). Complete enzyme is called holoenzyme. The sigma factor separates from the holoenzyme. The enzyme without factor sigma factor is called core enzyme.
Promoter:
ADVERTISEMENTS:
There is a definite sequence of bases on DNA called promoter to which the RNA polymerase binds. RNA polymerase recognise this promoter and then locally unwinds the DNA strands in order to gain access to the bases to be copied. In prokaryotes, the promoter is a sequence of six bases which in generally TATAAT or its slight variant.
This sequence is called Pribnow box. It lies 5-10 bases before the first base to be transcribed and is also called – 10 region. This region is called upstream region and is denoted by a minus sign (-).
Pribnow box orients RNA polymerase in such a way that transcription proceeds in 5′ → 3′ direction. In addition several promoters have a second important region to the left of Pribnow box which is a – 35 sequence. It is another six base sequence TTGACA. The large RNA polymerase enzyme covers – 35 sequence, Pribnow box and transcriptional start site.
These two conserved sequences called – 35 and – 10 regions are 17-19 nucleotides apart.
Elongation:
ADVERTISEMENTS:
After initiation, elongation of RNA chain occurs. New ribonucleotides are added one by one. It is called polymerization. During elongation RNA polymerase copies DNA sequence accurately. This is known as processivity. Elongation of RNA proceeds only in 5′ —> 3′ direction like DNA replication.
During elongation phase of transcription, the RNA strand base pairs temporarily with DNA template to form a short RNA-DNA hybrid double helix. Then the RNA separates and two strands of DNA again form duplex structure. DNA strands must unwind over a short distance to enable RNA polymerase to synthesize RNA strand complementary to one of DNA strands.
This unwinding of DNA helix opens up the DNA helix. A wave of unwinding is generated by RNA polymerase. In this way DNA is unwound ahead and rewound behind as RNA is transcribed.
RNA polymerase also performs proof reading functions as transcription is less accurate than replication. In replication one mistake is made for every 10,000,000 nucleotides added while in transcription one mistake is made for every 10000 nucleotides added.
Termination and Release:
ADVERTISEMENTS:
After the RNA synthesis is completed, RNA polymerase reaches a stop signal or a termination factor on DNA. In E. coli there are two types of termination signals, Rho-independent and Rho-dependent.
In Rho-independent termination is also called intrinsic termination. Near the 3′-end of newly synthesized RNA, there is a sequence of inverted repeats, about 20 nucleotides long, which form a self complementary hairpin or stem structure. At the end of this hairpin structure there is a sequence of uracil bases which are transcribed from adenine bases on the template strand. This A = U hybrid region has weakest bonds, and therefore is unstable and leads to the termination and dissociation of newly synthesized RNA molecule. This is self-terminating and depends upon the DNA base sequence.
Rho-dependent termination is dependent on a protein factor called Rho factor. Here Rho- protein has an ATP-dependent RNA-DNA helicase activity that causes release of newly synthesized RNA molecule.
In Prokaryotes Transcription and Translation Go On Simultaneously:
As prokaryotes do not have a nuclear membrane, transcription and translation occur in the same compartment. Even as the synthesis of RNA is in progress, ribosomes bind to the free 5′-end of mRNA and start protein synthesis or translation. The ribosomes bind at the free 5′-end mRNA while the 3′-end is still being transcribed. This is called coupled transcription and translation. Transcription, translation and degradation of mRNA all occur in the same 5′ → 3′ direction.
Transcription in Eukaryotes:
The basic features of transcription and structure of mRNA in eukaryotes are similar to those of bacteria. But there are certain differences between prokaryotes and eukaryotes. In eukaryotes, the mRNAs are generally monocistronic as compared to polycistronic mRNA of prokaryotes.
RNA Polymerase of Eukaryotes:
There are three kinds of RNA polymeases in eukaryotes. They are RNA polymerase 1, RNA polymerase II and RNA polymerase III.
RNA polymerase I transcribes only rRNAs i.e., 5.8S, 18S, and 28S, rRNAs. The RNA polymerase II transcribes all mRNAs. The RNA polymerase III transcribes tRNA and 5S rRNA.
Promoters for RNA Polymerase II:
The promoters for RNA polymerase II in eukaryotes are more complex. One promoter consists of a sequence of bases which lie – 25 bases upstream of transcription starting site. It consists of a sequence of seven bases TATAAAT called TATA box. It is also called Hogness box. It can be compared to Pribnow box of prokaryotes. Only TATA box is present in almost all the eukaryotes. In many cases another sequence is also present which lies – 75 base region called CAAT box. It has a sequence GGTCAATCT. Transcription start site lies in the initiator sequence where DNA is unwound.
Various transcription factors bind to TATA box and other sequences. RNA polymerase does not bind to these promoter sequences directly but to these factors as eukaryotic DNA is in the form of chromatin.
Post Synthesis Processing of RNA:
Newly synthesized RNA is called primary transcript or precursor RNA (pre-RNA). These RNA molecules undergo extensive changes called or processing to form mature RNA molecules which take part in protein synthesis. The most complex and characteristic feature of pre-mRNA is the presence of non-coding regions called introns present in between coding regions called exons.
These non-coding regions or introns are removed and discarded before the protein synthesis can take place. They are removed by a process called splicing. This is essential to get a correct sequence of amino acids of a polypeptide.
Almost all kinds of RNA molecules undergo post synthesis processing. Prokaryotic mRNA is generally not processed. Eukaryotic mRNA undergoes maximum processing. Both prokaryotic and eukaryotic tRNAs and rRNAs undergo extensive processing.
2006 Nobel Prize:
Roger Korenberg of America won Nobel Prize for Chemistry for describing the molecular basis of transcription in eukaryotic cells, how the information in the genes is copied and transferred.
His father Arthur Korenberg had won Nobel Prize in medicine in 1959 also for genetic work.
Summary:
Genes are expressed by transfer of genetic information from DNA to RNA. From RNA the information is used for synthesizing proteins. RNA is synthesized from a portion of one
strand of DNA which acts as a template. One gene or group of genes is transcribed at any one time producing one to numerous copies. On the other hand in replication both DNA strands are copied to produce two copies similar to the parent DNA molecule.
Polymerization is similar in both replication and transcription. Ribonucleotides of RNA are ATP, GTP, CTP, UTP. Thymine of DNA is replaced by uracil in RNA. RNA strand formed is complementary and antiparallel to the DNA template strand. Unlike replication, no primer is required in transcription. In prokaryotes a single RNA polymerase enzyme synthesizes all types of RNAs.
There is a definite sequence of bases on DNA called promoter on which the RNA polymerase binds. In prokaryotes a sequence of six bases called Pribnow box lies 5-10 bases before the transcriptional start site. Another sequence called – 35 sequence may be present. RNA polymerase binds these promoter sites and unwinds the DNA molecule for copying the portion of DNA template.
In E. coli two types of termination signals, Rho-independent and Rho- dependent signals are present. In prokaryotes, transcription and translation go on simultaneously. In eukaryotes basic steps of transcription are similar to prokaryotes. But some major differences are there. In eukaryotes three kinds of RNA polymerase enzymes are present. RNA polymerase I transcribes only rRNAs i.e. 5.8S, 18S and 28S rRNAs. The RNA polymerase II transcribes all mRNAs.
The RNA polymerase III transcribes tRNA and 5S rRNA. Promoters for RNA polymerase II are -25 sequence called TATA box and – 75 sequence called CAAT box. Newly synthesized RNA is called primary transcript or pre-RNA. These RNA molecules undergo extensive changes called processing to form mature RNA molecules which take part in protein synthesis. Processing occurs by splicing in which non coding regions called introns are spliced out and coding regions or exons are joined.