ADVERTISEMENTS:
In this article we will discuss about:- 1. Introduction to Protein Biosynthesis 2. Initiation of Protein Biosynthesis 3. Elongation 4. Termination 5. Polysomes.
Contents:
- Introduction to Protein Biosynthesis
- Initiation of Protein Biosynthesis
- Elongation of Protein Biosynthesis
- Termination of Protein Biosynthesis
- Polysomes
1. Introduction to Protein Biosynthesis:
ADVERTISEMENTS:
Protein biosynthesis can be explained by initiation, elongation and termination.
The mRNA nucleotide sequence is translated into the sequence of amino acids of the specified protein. The translation of the mRNA starts near its 5′-terminal with the formation of the corresponding amino terminal of the protein molecule.
In eukaryotic organisms, the process of transcription is a nuclear one; mRNA translation occurs in the cytoplasm. The simultaneous transcription and translation in eukaryotic organisms make the processing necessary to generate mature mRNA from the primary transcript -hnRNA.
2. Initiation of Protein Biosynthesis:
ADVERTISEMENTS:
The process of initiation needs tRNA, rRNA, mRNA, and at least 10 eukaryotic initiation factors (elFs), some of which have multiple (3 to 8) subunits. GTP, ATP and amino acids are also involved.
This process can be divided into four steps:
i. Dissociation of the Ribosome into its 40s and 60s Sub-units.
ii. Formation of the 40s Pre-initiation Complex.
iii. Formation of the 40s Initiation Complex.
iv. Formation of the 80s Initiation Complex.
i. Dissociation of the Ribosome:
(a) The binding of two eukaryotic initiation factors eIF-3 and elF-IA to the 40s subunit favours dissociation of the 80s ribosome into its 40s and 60s subunits and prevents reassociation.
(b) Reassociation is also prevented by the binding of eIF-3A to the 60s subunits.
ADVERTISEMENTS:
ii. Formation of the 40s Pre-initiation Complex:
(a) GTP binds eIF-2 and this binary complex then binds to met- tRNA, a tRNA is involved in binding to the initiation codon AUG.
(b) This above complex binds to the 40s ribosomal subunit to form the 40s pre-initiation complex which is stabilized by association with eIF-3 and elF-IA.
(c) The eIF-2 consists of α and β subunits. The eIF-2α is phosphorylated by a cAMP- independent protein kinase which is activated when a cell is under stress and also when the energy expenditure is deleterious. Amino acid and glucose starvation, virus infection, serum deprivation, and heat shock are included in such conditions.
ADVERTISEMENTS:
(d) The phosphorylated eIF-2α inactivates and binds tightly to the GTP-GDP recycling protein eIF-2β. This prevents formation of the 40s pre-initiation complex and blocks protein synthesis.
iii. Formation of the 40s Initiation Complex:
(a) The methyl-guanogyl triphosphate cap help’ the binding of mRNA to the 40s pre-initiation complex.
(b) A cap binding protein complex, eIF-4F, binds to the cap through one of its subunits. Then eIF-4A and eIF-4B bind and reduce the complex secondary structure of the 5′-end of the mRNA through their ATPase and helicase activities.
ADVERTISEMENTS:
(c) ATP hydrolysis is required in the association of mRNA with the 40s pre-initiation complex to form the 40s initiation complex.
(d) The eIF-3 is a key protein since it binds to mRNA and the 40s ribosomal subunit with high affinity.
(e) The eIF-4F (4F) complex is very important in controlling the rate of protein translation. 4F is a complex consisting of elF- 4E (4E), which binds to the m7G cap structure at the 5′-end of the mRNA, eIF-4A, which is an RNA-dependent ATPase that helps unwind the RNA, and p220. 4E is important since cap-binding is a rate-limiting step in translation. Overexpression of 4E causes malignant transformation of cells.
(f) The phosphorylation of 4E (on serine 53) and activation of the protein are caused by insulin and mitogenic growth factors.
ADVERTISEMENTS:
(g) A recently discovered family of proteins 4E-BP1 (and the closely related protein PHAS-1) and 4E-BP2 bind to and inactivate 4E. These proteins are again phosphorylated by insulin and growth factors via the stimulation of serine protein kinase resulting in the dissociation of the binding protein from 4E, the assembly of 4E and other components of the 4F complex in the mRNA cap, and initiation of protein synthesis.
iv. Formation of the 80s Initiation Complex:
(a) The hydrolysis of the GTP bound to eIF-2 by eIF-5 is caused by the binding of the 60s ribosomal subunit to the 40s initiation complex. This reaction promotes the release of the initiation factors bound to the 40s initiation complex (these factors are then recycled) and the rapid association of the 40s and 60s subunits to form the 80s ribosome.
(b) Ultimately, the met-tRNA on the P site of the ribosome becomes ready for the elongation cycle to start.
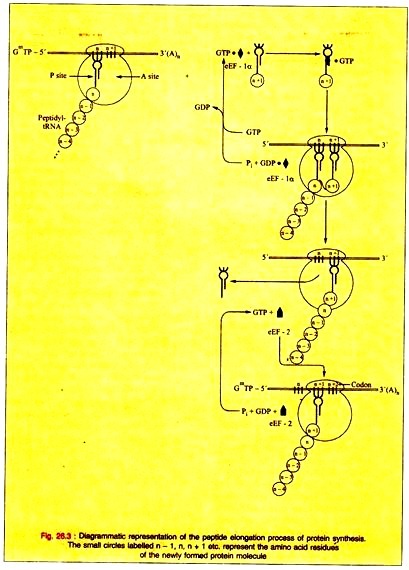
3. Elongation of Protein Biosynthesis:
ADVERTISEMENTS:
Elongation is a cyclic process consisting of several steps catalyzed by elongation factors (eEF).
The steps are:
i. Binding of Aminoacyl-tRNA to the A site.
ii. Peptide Formation.
iii. Translocation.
i. Binding of Aminoacyl-tRNA to the A site:
ADVERTISEMENTS:
The aminoacyl-tRNA binds to the A site of the complete 80s ribosome requiring proper codon recognition.
The complex formed by the elongation factor eEF-1α with GTP and the aminoacyl-tRNA enters the A site with the release of eEF-1 α-GDP and phosphate. The eEF-1 α-GDP then recycles to eEF-1 α-GTP by the help of other soluble protein factors and GTP.
ii. Peptide Bond Formation:
(a) The α-amino group of the new aminoacyl-tRNA in the A site has a nucleophilic attack on the esterified carboxyl group of the peptidyl-tRNA occupying the P site being catalyzed by a protein component peptidyltransferase of the 60s ribosomal subunit. This enzymic activity is performed by some RNA component of the ribosome, perhaps the 23s rRNA. This shows the direct role for RNA in protein synthesis.
(b) The amino acid on the aminoacyl-tRNA is already activated for which no further energy source is required for this reaction.
iii. Translocation:
(a) The discharged tRNA is quickly dissociated from the P site on the removal of the peptidyl moiety from the tRNA in the P site.
(b) The elongation factor 2 (eEF-2) and GTP are responsible for the translocation of the newly formed peptidyl-tRNA at the A site into the empty P site. GTP is hydrolyzed to GDP and phosphate during the translocation.
(c) The translocation of the newly formed peptidyl-tRNA frees the A site for another cycle of aminoacyl-tRNA codon recognition and elongation.
(d) ATP is hydrolyzed to an AMP which is equivalent to the hydrolysis of two ATPs to two ADPs and phosphates in the charging of the tRNA molecule with the aminoacyl moiety. GTP is also hydrolyzed to GDP during the entry of the aminoacyl- tRNA into the A site.
The hydrolysis of GTP to GDP and phosphate is caused by the translocation of the newly formed peptidyl-tRNA in the A site into the P site by eEF-2. The formation of one peptide bond causes the hydrolysis of two ATP molecules to ADP and two GTP molecules to GDP or the hydrolysis of four high-energy phosphate bonds. This process occurs rapidly.
(e) As many as six amino acids are incorporated per second in case of an eukaryotic ribosome and eighteen amino acids are incorporated per second in case of prokaryotic ribosome. Thus, the process of peptide synthesis is enhanced with great speed and accuracy until a termination codon is reached.
4. Termination of Protein Biosynthesis:
(a) The nonsense or the terminating codon of mRNA (UUA, UAG, UGA) appears in the A site after many cycles of elongation in the process of polymerization of the specific amino acids into a protein molecule.
(b) Normally there is no tRNA with an anti- codon to recognize such a termination signal.
(c) The existence of a termination signal in the A site is recognized by the releasing factors (eRF) which are proteins that hydrolyze the peptidyl-tRNA bond when a nonsense codon occupies the A site.
(d) The releasing factor enhances the hydrolysis of the bond between the peptide and the tRNA occupying the P site in conjugation with GTP and the peptidyl transferase. This hydrolysis caused by the addition of a water molecule releases the protein and the tRNA from the P site.
(e) The 80s ribosome dissociates into its 40s and 60s subunits upon hydrolysis and release. These subunits are recycled.
(f) The mRNA is then released from the ribosome which dissociates into its component 40s and 60s subunits and another cycle can be repeated.
The machinery of Protein synthesis can respond to environmental threats:
(a) Ionized iron (Fe++) is prevented from reaching toxic levels within cells by ferritin which is an iron binding protein.
(b) Ferritin synthesis is stimulated by elemental iron by causing the release of a cytoplasmic protein that binds to a specific region in the 5′ non-translated region of ferritin mRNA. The disruption of this protein -mRNA activates ferritin mRNA and results in its translation. This mechanism causes rapid control of the synthesis of a protein that resists Fe++, a toxic molecule.
(c) Some viral mRNAs are translated more efficiently than those of the host cell. Other viruses inhibit host cell protein synthesis by preventing the association of mRNA with the 40s ribosome.
The activity of many proteins is affected by posttranslational processing:
(a) The long polycistronic proteins are synthesized from one long mRNA molecule by some animal viruses and hepatitis A virus. These protein molecules are cleaved at specific sites to give the several specific proteins required for viral function.
(b) In animal cells, many proteins are synthesized from the mRNA template as a precursor molecule which is then modified to form the active protein. The prototype is insulin which is synthesized as a prohormone that folds to allow the disulfide bridges to form.
(c) Many other peptides are synthesized as pro-proteins that need modifications before having biologic activity. Many of the posttranslational modifications involve the removal of amino terminal amino acid residues by specific amino-peptidases.
The role of antibiotics in the inhibition of protein synthesis in bacteria:
ADVERTISEMENTS:
(a) The bacterial ribosome is smaller (70s rather than 80s) and has a complement of RNA and protein molecules. This difference is important for clinical purposes because many effective antibiotics interact with the proteins of prokaryotic ribosomes and thus inhibit protein synthesis.
This causes growth arrest or death of the bacterium. The most useful antibiotics e.g., tetracyclines, erythromycin and chloramphenicol, do not interact with components of eukaryotic ribosomal particles and hence are not toxic to eukaryotes.
(b) Other antibiotics e.g., puromycin inhibit protein synthesis on all ribosomes. Puromycin is structurally similar to tyrosinyl-tRNA. This drug is incorporated via the A site of the ribosome into the carboxy-terminal position of a peptide but causes the premature release of the polypeptide.
Puromycin effectively inhibits protein synthesis in both prokaryotes and eukaryotes. Cycloheximide inhibits peptidyltransferase in the 60s ribosomal subunit in eukaryotes by binding to an rRNA component.
(c) Diphtheria toxin catalyzes the ADP- ribosylation of eEF-2 in mammalian cells and specifically inhibits mammalian protein synthesis. Many animals (e.g., mice) are resistant to diphtheria toxin. This is due to the inability of diphtheria toxin to cross the cell membrane.
(d) Purotnycin and Cycloheximide are not clinically useful but are important in elucidating the role of protein synthesis in the regulation of metabolic processes.
5. Polysomes:
(a) Many ribosomes on the same mRNA molecule form a polyribosome or polysome.
(b) The mass of the mRNA molecule is quite small compared to the mass of even a single ribosome.
(c) A single mammalian ribosome can synthesize about 100 peptide bonds per minute.
(d) Polyribosomes actively synthesizing protein can exist as free particles in the cellular cytoplasm or may be attached to endoplasmic reticulum. This attachment is due to the rough appearance of the polyribosomes. Some of the proteins synthesized by the attached polyribosmes are packaged by the Golgi apparatus.
(e) The polyribosomal particles free in the cytosol are responsible for the synthesis of proteins required for intracellular functions.
The energy requirements for the formation of one peptide bond:
i. 2 ATP (charging of amino acids).
ii. 2 GTP (one for transferring amino acyl tRNA and another for transferring tRNA from A site to P site).