ADVERTISEMENTS:
In this article we will discuss about the representation of gene for gene interaction in resistance development.
Plants, in nature, contain several resistance genes as a defensive role. Since the turn of this century plant breeders have used these disease resistance genes commonly known as ‘R’ genes to counter plant disease. Knowledge about genetic and biochemical basis of plant disease resistance has been found to be accumulated considerably.
Plant breeders now have clear idea that disease resistance is controlled by Mendelian genes. During evolution these genes originated along with the pathogen in co-evolved fashion. This outstanding discovery accomplished by plant breeders has led to the use of interspecific cross between crops and their wild relative as source of resistance germplasm.
ADVERTISEMENTS:
A molecular cloning and characterization of ‘R’ genes have been analysed after 1992. In the mechanism of disease resistance, there are specific ‘R’ genes presence in the plant host and corresponding virulence genes in the pathogen. Although nature containing vast array of potential pathogen, only few plants exhibit resistance. Plant exhibit resistance to pathogens in number of ways.
They are hypersensitive response (HR), localized induce cell death in the host plant at the site of infection. Hypersensitive responses are prominently checking the growth of pathogen. Genetic basis of HR-mediated resistance has been recorded by flor. He proved that disease resistance of flax to the fungal pathogen Melampsora lini is due to the interaction between pair of genes in the host and the pathogen.
His work provides theoretical basis for molecular cloning of pathogen avirulence (avr) genes and their corresponding plant R genes. In the gene for gene interaction during plant defence, avirulence (avr) genes of pathogen are expressed to produce elicitors (Fig. 20.1).
These elicitors are signal molecules recognised by host plant and leads to the cascade of hypersensitive reaction. The R genes of host plants are presumed to encode receptors for these elicitors. The elicitor signal molecule triggers the expression of a cascade of host genes that led to hypersensitive reaction and check pathogen growth.
Gene for gene interaction pathway shows involvement of signal transduction. Some of the hypersensitive reaction (HR) which is common to different pathogen include rapid oxidative burst, ion flux by K+H+ exchange, cellular decomposition, cross linkage and strengthening of cell wall. In addition, induction of pathogenesis related (PR) proteins takes place in response to pathogen attack.
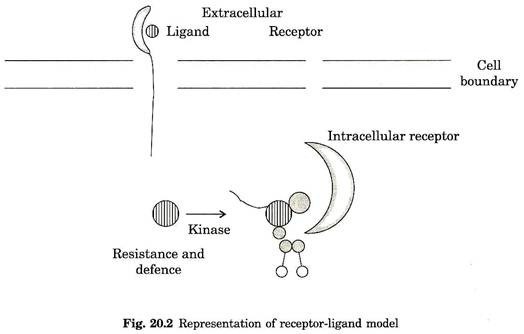
Several crop plants such as tomato, bean, potato and barley have been used to study crop responses to pathogen attack. Based on the receptor signal model it was postulated that pathogen avr gene specify elicitor signal molecule induce disease resistance in host plant that contain ‘R’ genes (Fig. 20.2).
In one of the case study, the interaction between leaf mold pathogen Cladosporium fulvum and tomato plant have been well established. The two avirulence genes, avr9 and avr4, belongs to C. fulvum encode precursor of elicitor peptide that specifically induce HR response in plant that encodes corresponding ‘R’ genes Cf-9 and Cf-4 respectively.
In another confirmed case, the avirulence gene avr D of Pseudomonas syringae encodes enzymes involved in the synthesis of syringolids that elicit HR in soyabean carrying R gene Rpg4. Similarly, TMV coat protein functions as intracellular elicitor that induce HR in Nicotiana sylvestris carry corresponding gene known as N, these studies reveals that having a avirulence has definite advantage to pathogen and that can enhance virulence on host (susceptible) that do not carry ‘R’ genes.
Gene for Gene Relationship:
Several R genes have been isolated and characterized by positional cloning and transposon tagging. The classic gene for gene relationship was characterized in tomato pto gene. The pto gene confers resistance to P. syringae (pst) carrying avirulence gene avr pto.
Expressed product of pto gene confirmed that it encodes a serine-threonine protein kinase. This enzyme probably involved in signal transduction. The fine locus of PTO consists of cluster of five to seven genes, all homologous to PTO. Some of these genes like FEN, PRF and others conferred resistance and sensitivity to organophosphorus insecticide fenthion.
In another case study, four ‘R’ genes have been cloned to confirm the concept of gene-for- gene relation, for example Arabidopsis carrying over RPS2 genes, which confer resistance to pathogen P. syringae carries avr RPS2. The lack of RPS2 ‘R’ gene in mutant Arabidopsis result in complete absense of HR during pathogen (P. syringae) attack carries RPt2.
ADVERTISEMENTS:
The RPS2 gene was then cloned by map-based strategy to initiate HR process. Transposon tagging helps in the identification of several resistance gene like flax L6, tobacco N and tomato cf-9 gene. Translation product of RPS2, N, cf-9 and L6 R genes revealed that their proteins contain leucine, rich repeat (LRR).
These four genes are different from both maize HM, and the tomato pto resistance (R) genes. RPS2, N, and L6 except cf-9 sharing some homology and contain conserved nucleotide binding site (NBS). The sequence of rice Xa21 R gene, which confers resistance to Xanthomonas oryzae race 6, predicted that their protein also conferring both leucine rich repeat motif and serine-threonine-kinase like domain suggested a role in cell surface recognition of pathogen ligand and subsequent activation of an intracellular defence response characterization of Xa21 gene lead to engineered resistance in rice.
Further studies on ‘R’ proteins revealed that Cf9 protein appears to consist of primarly of extra cytoplasmic LRR and also suggested that Cf-9 is a receptor for extracellular ligand provided by the avr 9 elicitor peptide of pathogen. Another R protein RPS2 contains leucine zipper at NH2-terminus, probably involved in protein dimerization.
The isolation of over 30 plant disease resistance genes revealed that most genes encode putatively cytoplasmic proteins with a nucleotide binding site (NBS) and a leucine-rich repeat (LRR). However, the recent isolation of the Arabidopsis RRSI-R gene has NBS-LRR subtype that contains αC-terminal extension with a putative nuclear localization signal and a DNA binding domain.
ADVERTISEMENTS:
Deviation in gene-for-gene resistance has also been established for example, HMI gene in maize, which control resistance to Cochliobolus carbonum, HMI encodes NADPH dependent HC toxin reductase, which inactivate HC toxin, pathogenecity factors, produced by fungus C. carbonum. The genetic interaction between maize and pathogen fungus different from gene for gene interaction because toxin-deficient C. carbonum fail to impose disease in maize that do not carry HMI.