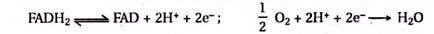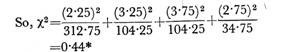ADVERTISEMENTS:
Let us make an in-depth study of the biomaterial-nanoparticle hybrid systems. The below given article will help you to learn about the following things:- 1. Introduction to Biomaterial-Nanoparticle 2. The Synthesis and Properties of Biomaterial-Functionalized Nanoparticles 3. Biomaterial-Functionalized Nanoparticles for Controlled Chemical Reactivity 4. Aggregation of Biomaterial-Functionalized Nanoparticles and Others.
Introduction to Biomaterial-Nanoparticle:
The unique electronic, optical and catalytic properties of metal and semiconductor nanoparticles (1-200 nm), together with the different methods available for the preparation of nanoparticles of controlled shape and size, provide exciting building blocks for Nano scale assemblies, structures and devices. Nobel Laureate Richard Feynman in his visionary lecture ‘There is plenty of room at the bottom’ inspired the concepts for the rapidly exploding research topic of nanotechnology.
Although the term ‘nanotechnology’ had not appeared on the horizon, Feynman said: ‘What I want to talk about is the problem of manipulating and controlling things on a small scale… What I have demonstrated is that there is room—that you can decrease the size of things in a practical way… I will not discuss how we are going to do it, but only what is possible in principle… We are not doing it simply because we haven’t yet gotten around to it.’
ADVERTISEMENTS:
Four decades later scientists have learnt that the manipulation of atoms, molecules and clusters on surfaces is feasible, and that new fundamental physics governs the properties of Nano-objects. The miniaturization of structures by conventional and electron-beam lithography is reaching the theoretical limits of ca. 50 nm. For the further miniaturization of chemical objects, alternative approaches must be developed.
Following Feynman’s vision, one may employ atoms and molecules as building units for the ‘bottom-up’ assembly and construction of architectures with nanometer dimensions. Nanoparticles consisting of metals (e.g. Au, Ag, Pt, and Cu) or semiconductors (e.g. PbS, Ag2S, CdS, CdSe, and TiO2 seem to be attractive units for the engineering of such structures. The unique electrical charging properties of these particles, as well as their optical and photo physical features, such as size-controlled Plasmon absorbance, photonic electron-hole pair generation, and fluorescence, allow the addressing of particles by external electronic and photonic signals.
A variety of synthetic methodologies for the preparation of nanoparticles within a narrow size distribution are available. Often, the nanoparticles are prepared by ‘wet chemistry’ procedures, where the clustering of the metal atoms or semiconductor molecules proceeds in the presence of a surface capping ligand. This capping ligand binds to the metal/semi-conductor clusters, prevents aggregation of the particles into bulk material, and controls the final dimensions of the nanoparticles.
Many capping systems are available, including hydrophobic monolayers, positively or negatively charged monolayers, and polymer layers. Association of molecular units to the nanoparticles introduces chemical functionalities that can provide recognition or affinity interactions between different appropriately modified particles, and thereby dictate structure when aggregation occurs.
ADVERTISEMENTS:
New collective properties of aggregated particles such as coupled Plasmon absorbance, inter-particle energy transfer, and electron transfer or conductivity may be observed in the clustered assemblies. The chemical functionalities associated with nanoparticles enable the assembly of 2D and 3D nanoparticle architectures on surfaces.
Composite layered or aggregated structured structures of molecule or macro-molecule-cross-linked nanoparticles on surfaces have been prepared, and the specific sensing of substrates, tunable electroluminescence, and enhanced photo electrochemistry have been accomplished.
The assembly of nanoparticle architectures on surfaces has also led to the fabrication of Nano scale devices such as single electron transistors, nanoparticle-based molecular switches, metal-insulator-nanoparticle-insulator metal (MINIM) capacitors, and others.
Several reviews have addressed recent advances in the synthesis and properties of nanoparticles and the progress in the integration of composite nanoparticle systems with surfaces. The conjugation of nanoparticles with biomaterials is a tempting research project that provides a route into Nano biotechnology.
Evolution has optimized fascinating macromolecular structures exhibiting unique recognition, transport, and catalytic properties. The conjugation of nanoparticles with biomaterials could provide electronic or optical transduction of the biological phenomena. Enzymes, antigens and antibodies, and receptors have dimensions in the range of 2-100 nm, comparable to those of nanoparticles, and thus the two have structural compatibility.
Several fundamental features show biomaterials to be important future building blocks for nanoparticle architectures:
1. Biomaterials reveal specific and strong complementary recognition interactions, e.g., antigen-antibody, nucleic-acid-DNA, hormone-receptor. The functionalization of a single kind of nanoparticle or different kinds of nanoparticles with biomaterials could lead to biomaterial-nanoparticle recognition and thus to self-assembly.
2. Various biomaterials include several binding sites, e.g., the two Fab-sites of antibodies, the four binding domains of streptavidin or concanavalin A. This allows the multidirectional growth of nanoparticle structures.
3. Proteins may be genetically engineered and modified with specific anchoring groups. This facilitates the aligned binding to nanoparticles, or the site-specific linkage of the biomaterial to surfaces. Consequently, directional growth of nanoparticle structures may be dictated. Furthermore, other biomaterials, such as double-stranded DNA, may be synthetically prepared in complex rigidified structures that act as templates for the assembly of nanoparticles by intercalation, electrostatic binding to phosphate groups, or association to functionalities tethered to the DNA.
ADVERTISEMENTS:
4. Enzymes provide catalytic tools for the manipulation of biomaterials. For example, the ligation of nucleic acids or the endonuclease scission processes of nucleic acids provide effective tools for controlling the shape and structure of biomaterial-nanoparticle hybrid systems. In this context, it is important to note that Mother Nature has developed unique bio-catalytic replication processes. The use of biocatalysts for the replication of biomaterial-nanoparticle conjugates may provide an effective system for the formation of nanostructures of predesigned shapes and compositions.
It is the aim of this article to review the synthesis of biomaterial-nanoparticle hybrid systems, as well as the organization of these systems as functional devices (Fig. 9.1). We describe their properties, the methods of assembling two dimensional and three-dimensional nanoparticle structures on surfaces, and the operation of these biomaterial-nanoparticle structures as functional devices. It is our aim to highlight the fact that the scientific accomplishments of the past few years have already addressed important aspects of Nano biotechnology.
The Synthesis and Properties of Biomaterial-Functionalized Nanoparticles:
Biological molecules have been immobilized on polymer matrices and inorganic supports by a variety of techniques, including physical adsorption, electrostatic binding, specific recognition and covalent coupling. These supports, modified with biological molecules such as proteins/enzymes, antigens/antibodies, and DNA/oligonucleotides, have been used for numerous biotechnological applications, including affinity separations, bio sensing, bioreactors, and biofuel cells. Recently, these immobilization techniques—developed for the functionalization of macro-size supports have been applied to bring together biomolecules and nanoparticles.
Functionalization by Electrostatic Adsorption:
ADVERTISEMENTS:
The simple adsorption of biomolecules on nanoparticles has frequently been used and studied. These include biomolecules ranging from low-molecular-weight organic substances (e.g. Vitamin C) to large protein/enzyme molecules. In the case of nanoparticles that are stabilized by anionic ligands such as carboxylic acids (citrate, tartarate, lipoic acid), the adsorption of proteins originates from electrostatic interactions. For example, gold and silver nanoparticles produced by citrate reduction have been functionalized with immunoglobulin (IgG) molecules at pH values slightly above the isoelectric point of the citrate ligand.
This allows effective binding between the positively charged amino acid side chains of the protein and the negatively charged citrate groups of the colloids. Other examples of protein coating through electrostatic interactions include the direct adsorption of heme-containing redox enzymes at citrate-stabilized silver nanoparticles and the binding of basic leucine zipper proteins to lipoid acid-stabilized semiconductor CdSecore/ZnSshell particles.
Differential scanning calorimetry (DSC) has been used to demonstrate the electrostatic nature of proteins adsorbed on silica particles, and allows the calculation of thermodynamic parameters. The electrostatic deposition of biomolecules, particularly proteins or enzymes, can also be extended to a multi-layer level. Protein molecules electrostatically attracted to the charged nanoparticles can provide a base interface for the further deposition of an oppositely charged polyelectrolyte polymer that again allows the deposition of a secondary protein layer.
Multilayer films of bovine serum albumin (BSA), immunoglobulin G (IgG), b-glucosidase (b- GLS), and glucose oxidase (GOx), urease (Ur), and horseradish peroxidase (HRP) have thus been assembled on polystyrene nanoparticles by the alternate deposition of the protein and oppositely charged polymers.
ADVERTISEMENTS:
The protein/polymer multilayer shell could be varied from several to hundreds of nanometers in thickness. This strategy permits the preparation of functional films with a high density of enzyme molecules on nanoparticles. Similarly, the electrostatically driven adsorption of negatively charged DNA molecules on positively charged CdS nanoparticles has been studied in detail.
An increase in the loading of nanoparticles of organic materials, upon layer-by- layer deposition, increases the sensitivity of analytical methods when these particles are utilized. For example, polystyrene particles were functionalized with a multilayer assembly of polyelectrolytes functionalized with fluorescent dye.
The assembly was terminated with an IgG layer that allowed the use of the modified particles as labels in fluorescence immuno analysis. The increase of the loading of the particles with the dye because of multilayer deposition resulted in higher fluorescence output, and thus in a higher sensitivity of the analysis.
Functionalization by Chemisorption of Thiol-Derivatized Biomaterials:
In some cases, strong chemisorption of proteins on Au nanoparticles can originate from the binding of thiol groups (from cysteine residue) existing in the proteins [e.g. immunoglobulin’s (IgG), serum albumin] to the Au surface (Fig. 9.2 A). If no such residues are available in the native proteins, thiol groups can be incorporated by chemical means [e.g., with Traut’s reagent, 2-iminothiolane (1) in Fig. 9.2 B] or by genetic engineering.
ADVERTISEMENTS:
Some proteins and enzymes preserve their native structures and activities when they are directly adsorbed on nanoparticles. For instance, a surface-enhanced resonance Raman (SERR) spectrum of native hemoglobin was obtained when it was adsorbed on citrate-stabilized Ag colloids. Reversible di-oxygen and carbon monoxide binding and the R to T-state transition were also observed.
A non-heme iron enzyme, chlorocatechol di-oxygenase, physically adsorbed on citrate-reduced Ad colloid, also showed its native structure and bio-catalytic activity upon application of SERR spectroscopy. However, physically adsorbed biomolecules can be readily lost from the surface, and adsorbed proteins are often prone to denaturation, thereby losing their bio-catalytic or bio-recognition activities.
Careful investigations of some proteins and enzymes directly adsorbed on nanoparticles demonstrate their conformational changes and loss of biological activity. For example, circular dichroism (CD), DSC, and fluorescence spectroscopy applied to bovine fibrinogen adsorbed on TiO2 nanoparticles indicate that the a-helix content of the protein decreases markedly upon their adsorption.
Nucleic acids can be synthesized with tethered alkanethiol groups by using appropriate phosphoramidate precursors in a solid-phase synthesis. The nalkyethiolated nucleic acids have been used extensively in the preparation of DNA-functionalized gold nanoparticles and CdSecore/Znshell or CdS semiconductor nanoparticles.
DNA molecules functionalized with a steroid-disulfide derivative can readily bind to Au nanoparticles and demonstrate higher adsorption stability because two sulfur atoms of the disulfide anchor group are involved in the attachment process.
ADVERTISEMENTS:
Enhanced binding of nucleic acids to metal nanoparticles may be accomplished by the use of oligonucleotides that include several adenosyl phosphothiolate residues at their ends, thus enabling multi-point attachment to the nanoparticles.
Functionalization by Specific Interactions:
Nanoparticles functionalized with groups that provide sites for the affinity binding of biomolecules have been used for the specific attachment of proteins and oligonucleotides. For example, streptavidin-functionalized Au nanoparticles have been used for the affinity binding of biotinylated proteins (e.g. immunoglobulin’s and serum albumins) or biotinylated oligonucleotides. Likewise, biotinylated CdSe/CdS/SiO2 nanoparticles can be bound to streptavidin. Also, nanoparticle-antibody conjugates were used for affinity binding of their respective antigens. This may be advantageous, as antibodies associated with nanoparticles can demonstrate association constants with respective antigens that are even higher than those of the free antibody.
By covalently attaching proteins to nanoparticle surfaces, problems of instability, reversibility and inactivation can be overcome. Low-molecular bio-functional linkers that have anchor groups for attachment to nanoparticle surfaces and functional groups for further covalent coupling to the target biomolecules were extensively used for generating covalent conjugates of biomolecules and various Nano-particles.
Anchor groups such as thiols, disulfides, or phosphane ligands are often used for the binding of the bio-functional linkers to Au, Ag, CdS, and CdSe nanoparticles. These anchor groups readily substitute weakly adsorbed molecules stabilizing the nanoparticles, or may be incorporated in the nanoparticle synthesis, resulting in a nanoparticle surface providing functional groups for further reactions.
Alkoxysilane or halogen-silane groups are used for the covalent attachment of bio-functional linkers to the surfaces of Si02 and other oxide-coated nanoparticles. A wide variety of terminal functional groups is available in different bio-functional linkers: amino, active ester, and maleimide groups are used for the covalent coupling of biological compounds by means of carbodiimide-mediated esterification and amidation, or reaction with thiol groups. Single functional groups (e.g. active ester or amino units) on Au nanoparticles are available either by a statistical approach or by the modification of chemically defined clusters. These structures provide unique synthetic routes for the covalent binding of a single target biomolecule per nanoparticle.
Properties of Nanoparticle-Biomaterial Composites:
The functionalization of nanoparticles with biomolecules results in changes in the properties of the nanoparticles and their interactions with the environment. Upon adsorption of vitamin C on TiO2 nanoparticles, the optical properties of the particles were red shifted by 1.6 e.V. This is the result of charge transfer, and originates from the specific binding of the electron-donating modifier to corner defects on the surface of the nanoparticles.
ADVERTISEMENTS:
The solubility of nanoparticles in water can be greatly improved by the functionalization of their surfaces with highly hydrophilic biomolecules. While Au nanoparticles modified with long chain alkanethiols are only soluble in low-polarity organic solvents, Au nanoparticles with w-carboxylic acid-functionalized alkanethiolate ligand shells are soluble in polar organic solvents such as ethanol and acetone.
Au nanoparticles modified with biomolecules such as thiopronin or coenzyme A demonstrates excellent solubility in water. Alteration of the chemical properties of the biomolecules covering nanoparticles by external signals (e.g., electrical, optical) can be used to control interactions of the modified nanoparticles with the environment, for example, to control the binding of a secondary modifier or the aggregation of the nanoparticles.
Complex formation between a diaminopyridine derivative and a flavin derivative was studied at an Au nanoparticle surface. It has been shown that more stable hydrogen bonding is provided when the flavin derivative is electrochemically reduced. This allows electrochemically controlled switching of the binding of the bioorganic molecules to the organic-functionalized shell of the nanoparticle. The interactions of biomaterial-functionalized nanoparticles with other functionalized nanoparticles or other biomaterials such as proteins or DNA molecules provided by affinity properties of the biomaterial shell components are described in the following sections.
Biomaterial-Functionalized Nanoparticles for Controlled Chemical Reactivity:
The interactions of functionalized nanoparticles with biomaterials or within biomaterial structures can control the chemical reactivity of the biomolecules. Alternatively, they can report on the state of the reaction or reactants, allowing control to be exercised externally.
It is well known that small molecules and polymers can affect the chemical reactivity of biomolecules. If there are several possible parallel reactions, the effect produced by a promoter/inhibitor on a specific chemical reaction can change the effective chemical path of the whole process, resulting in a regulation of the biochemical system.
Molecular labels (e.g., fluorescent dyes) incorporated into biomolecules can report on the state of the biomolecule, transducing the molecular operation into an output signal used, for example, in biomaterial-based diagnostics. Functionalized nanoparticles can operate in the same way, demonstrating properties of a bio-promoter/bio-inhibitor or a reporter, with some advantage over their molecular-sized counterparts due to the unique photo-physical and electronic properties of the nanoparticles.
Controlling DNA Reactivity:
Bio-catalytic reactions occurring at DNA molecules, such as transcription or translation, require the association of enzymes with the DNA chain. The competitive association of other molecules to the DNA chain can decrease the enzyme binding and thus inhibit the process, finally resulting in gene regulation.
It has been shown that small molecules, dendrites, and polymers are capable of efficient binding to DNA that in some cases leads to inhibition of the transcription machinery. In these systems, however, the creation of suitably pre-organized scaffolds for the controlled binding to DNA presents a significant design challenge.
For small molecules, this challenge arises from the detailed synthesis required for a rigid scaffold containing several functional groups. For polymer systems, the precise placements of substituents along the backbone as well as the control of the polymer macro-conformation in solution are difficult to achieve. An excellent solution to these problems is the application of ‘mixed monolayer-protected Au clusters’ (MMPCs).
The self-assembled monolayer coating of the Au nanoparticles presents a highly organized surface for the recognition of DNA that is of a similar size scale (6-10 nm) to that of DNA- binding proteins. The central metal core rigidities the particle, limiting the organic components to a much smaller subset of structures than a similarly sized polymer.
The mobility of the thiols on the nanoparticle surface also allows the formation of recognition elements on the nanoparticle surface to be controlled in subsequent steps. Nanoparticles for the recognition of double-stranded (ds) DNA were prepared by the partial displacement of octanethiol on the Au surface with 11-mercaptoundecyltrimethylammonium.
The mixed monolayer of this MMPC system is designed to interact with DNA based on the electrostatic attraction of the positively charged quaternary ammonium salt and the negatively charged DNA phosphate groups. These MMPCs bind to the ds-DNA in competition with the enzymes involved in gene regulation, thus resulting in effective inhibition of the bio-catalytic transcription process.
Reporting on DNA Reactivity:
DNA molecules functionalized at specific positions with a fluorescent dye and a quencher can produce a fluorescence output controlled by the distance separating the dye and the quencher, thus reporting on the conformation of the DNA chain. Au nanoparticles can quench fluorescence of chromospheres to an extent that is hundred fold better than molecular quenchers, and they also have higher quenching efficiency for dyes emitting in the near infrared region.
A DNA oligonucleotide was functionalized with a fluorescent dye at the 30 end, and an Au nanoparticle functionalized with a single maleimide group was covalently linked to the 50 end through a (CH2)6-SH group. The molecule could adopt two conformations (Fig. 9.3): a stem-loop structure with the fluorophore and the Au nanoparticle held in close proximity (hairpin state), and a rod-like structure with them far apart (open state).
The hairpin state (2) is self-generated because of the intra-molecular complementarity of the terminal parts of the DNA chain that make a ds-structure within the same-molecule, whereas the open state (3) is produced upon interaction and hybridization with an analyte DNA molecule (4) that exhibits complementary to the central part of the DNA chain.
This target DNA hybridizes with the modified DNA, resulting in the formation of ds-DNA, thus opening the hairpin, increasing the distance between the fluorophore and the nanoparticle, and thereby enhancing the fluorescence. The process and the reporting signal only occur in the case of complementarity between the analyte DNA and the central part of the sensing DNA. The system was thus successfully applied for single-mismatch detection in DNA sequences.
Aggregation of Biomaterial-Functionalized Nanoparticles:
The organization and patterning of inorganic nanoparticles into two and three dimensional (2D and 3D) functional structures is a fundamental prerequisite for the assembly of chemical, optical, magnetic and electronic devices.
Many approaches have been described for the formation of 2D and 3D arrays of metal and semiconductor nanoparticles, for example, using:
(i) Solvent evaporation of hydrophobic colloids,
(ii) Random inclusion of the nanoparticles into gels and glassy matrices,
(iii) Template-directed synthesis at structured surfaces in porous protein crystals or bacterial superstructures, and
(iv) Chemical coupling in solution by means of bivalent cross-linker molecules.
An example of the latter is the alkanedithiol-directed aggregation of Au nanoparticles. The aggregation of nanoparticles induced by specific biological interactions attracts interest as a self-assembly process for the building of complex nanostructures exhibiting new collective properties.
There are several advantages in utilizing biomaterials as building blocks of nanoparticle structures:
1. The diversity of biomaterials facilitates the selection of building units of predesigned size, shape and functionality.
2. The availability of chemical and biological means to modify and synthesize biomaterials, e.g. synthesis of nucleic acids of predesigned composition and shape, eliciting monoclonal antibodies, or modifying proteins by genetic engineering, pave the way to the construction of biomaterials for the assembly of nanoparticles.
3. Enzymes may act as bio-catalytic tools for the manipulation of the biomaterials.
4. Hydrolysis of proteins, scission or ligation of DNA or replication of nucleic acids may be employed as assembler tools of nanoparticle architecture through the manipulation of the biomaterial.
5. Mother Nature has developed routes for the repair of biomaterials. These processes may be applied for the stabilization of the biomaterial/nanoparticle structures.
6. Crosslinking nanoparticles with enzyme units may generate bio-catalytic assemblies of pre-designed functionality.
These different features of the biomaterial crosslinking units provide the flexibility for the generation of nanoparticle structures of tunable physical, chemical and functional properties. For the generation of biomaterial-cross-linked nanoparticles, two kinds of biomaterial-functionalized nanoparticles with complementary units should participate in the assembly process.
Biomaterials utilized in the fabrication of such biomaterial-Nano-particle aggregates include biological protein host-guest pairs such as biotin-avidin, hapten-antibody and complementary oligonucleotides.
Receptor-Induced Aggregation of Guest-Functionalized Nanoparticles:
Protein-based recognition systems can be used to organize inorganic nanoparticles into network-aggregates, for instance with the interaction between D-biotin and the biotin-binding protein streptavidin (Sav). The recognition between water-soluble biotin and the homotetrameric protein Sav is characterized by an extraordinary affinity constant of Ka > 1014 M-1, which makes it the strongest ligand-receptor binding interaction presently known. Another great advantage of Sav is its extreme chemical and thermal stability.
The applicability of the biotin-Sav system for generating supra-molecular aggregates is enhanced by the availability of various biotin analogs and recombinant Sav mutants. A wide range of rate and equilibrium constants (Ka from 100 to 1015 M-1) are available, allowing the construction of carefully designed nanoparticle aggregates. Many biotinylated materials are commercially available or can be prepared under mild conditions.
Two Routes for Nanoparticle Aggregation by Biotin Sav Interactions are Possible:
(i) Nanoparticles functionalized with biotin groups can be cross-linked with the tetra-valent Sav receptor and
(ii) Nanoparticles functionalized with Sav can be cross-linked with a dibiotin derivative.
Several synthetic routes can be applied to functionalize metal and semi-conductive
nanoparticles with biotin derivatives. In the simplest method, thiol or disulfide biotin derivatives are directly adsorbed on metal (e.g. Au, Ag) nanoparticles.
Alternatively, nanoparticles can first be coated with an organic ‘shell’ (e.g. by the polymerization of a trialkoxysilyl derivative or by polymer adsorption) and then covalently modified with biotin (e.g. by carbodiimide coupling). Au nanoparticles functionalized with adsorbed disulfide-derivatized biotin units and then cross-linked with the added Sav have been shown to yield an aggregate of Au nanoparticles with the biotin-Sav recognition pairs between the nanoparticles.
A similar architecture can be built by reversing the steps: Sav was interacted with the disulfide biotin derivative to produce a complex, which was then reacted with Au nanoparticles. In both cases, fast, spontaneous aggregation of the Au nanoparticles was observed, resulting in a non-ordered network of particles.
Dynamic light scattering, small-angle X-ray scattering and transmission electron microscopy were used to follow the aggregation process and to characterize the product. The kinetics of this process was fitted to the theoretical Smoluchowski model of aggregation, modified for nanoparticles. This kinetic analysis allows control of the aggregate structure by optimization of the conditions (molar ratio of the biotin-functionalized nanoparticles and the Sav-cross linker).
Another example of biotin-Sav-cross-linked nanoparticles involves the native protein ferritin, which consists of a hollow polypeptide shell, 8 and 12 nm in internal and external diameters, respectively, and a 5 nm diameter ferric oxide core.
The ferric oxide nanoparticle can easily be removed from the protein shell by reductive dissolution, and the empty cage of Apo ferritin can be re-mineralized in vitro with a range of inorganic oxides, sulfides or selenides (e.g. MnO, FeS, CdS, CdSe). A succinimidyl active ester derivative of biotin was covalently linked to the amino groups of 60-70 lysine residues provided by the polypeptide shell of ferritin, yielding a biotin-functionalized protein shell containing the ferric oxide core (Fig. 9.4).
Addition of Sav resulted in the crosslinking and therefore the aggregation of the functionalized ferric oxide nanoparticles. Considering the library of core nanoparticles possible, this ferritin-based aggregate is a very promising material for producing electronic, magneto- electronic, and optoelectronic devices.
Nanoparticle aggregates have also been produced by means of the interaction between antigens and antibodies. In the simplest approach, anti-DNP IgE (DNP, dinitrophenyl) antibodies were adsorbed on 12 nm Au nanoparticles and were then cross-linked with a synthetic antigen (bis-N-2,4-dinitrophenyloctamethylene diamine) comprising two hapten DNP head groups separated by a spacer.
A more sophisticated approach allows the association of two different kinds of nanoparticles. For this purpose, two different nanoparticles functionalized with different kinds of bio-receptors (e.g. different antibodies or an antibody and Sav) should be cross-linked with a ligand containing two different head groups providing affinity binding to the two respective receptors. For example, two kinds of Au nanoparticles were functionalized, one with anti-DNP IgE and another with Sav.
These were cross-linked with a linker containing a biotin unit and a DNP group separated with a spacer. Each ligand recognizes its respective receptor (biotin binds to Sav, DNP binds to anti-DNP IgE), thus creating a network composed of both nanoparticles. A novel approach to nanoparticle crosslinking involves the application of specially designed de novo proteins with specific affinities to inorganic crystal surfaces. These structures can specifically bind directly to unmodified surfaces of various semi-conductive nanoparticles, thus resulting in their bridging and aggregation.
Based on a combinatorial library of about 109 random 12-mer peptides, de novo proteins with specific binding capabilities to single- crystal semiconductors including GaAs(100), GaAs(111), InP(100), and Si(100) have been discovered.
Nucleic Acid-Functionalized Nanoparticles for Controlled Aggregation:
DNA molecules are very powerful and versatile linkers for the controlled aggregation of nanoparticles. DNA chains are of a controllable length and composition, providing predictable properties and enormous control over complementarity. Ds-DNA provides rigid spacers that allow control over the distance between nanoparticles. Long DNA chains can be utilized as templates ready to accommodate complementary-DNA- functionalized nanoparticles in a controlled order and with controlled distances between the nanoparticles.
In order to demonstrate the DNA-mediated binding of nanoparticles, two batches of 13 nm diameter Au nanoparticles were separately functionalized with two thiol-derivatized non-complementary DNA oligonucleotides: 30-thiol-TACCGTTG-50 adsorbed on nanoparticle A and 50-AGTCGTTT-30-thiol adsorbed on nanoparticle B. A DNA linker was constructed composed of ds-DNA in the center and two single-stranded fragments on the ends. These single-stranded sections were complementary to the oligonucleotides connected to the nanoparticles. Addition of the linker to a mixed solution of the two DNA- functionalized Au nanoparticles results in crosslinking of the nanoparticles.
The resulting nanoparticle aggregate should theoretically have some degree of regularity, but this could not be observed, as both nanoparticles had identical cores. Aggregation changes the colour of the solution because of a collective phenomenon originating from the short-distance inter-particle separation.
Separate Au nanoparticles have a red colour as a consequence of their Plasmon absorbance, but when they are aggregated additional absorbance arise at lower energies because of inter-particle Plasmon coupling. This colour change can be used as a simple test for the recognition between complementary DNA sequences associated with the linker and the Au nanoparticles.
The association process is temperature dependent, and the aggregated Au nanoparticles can dissociate upon elevation of temperature and associate again upon decreasing the temperature, thus resulting in the reversible changes of the spectrum. In a control experiment, a non-complementary linker was added to the mixed particle solution, and no colour changes were observed.
In a more sophisticated investigation, a mixture of DNA-functionalized Au nanoparticles of diameters 40 nm (A) and 5 nm (B) was cross-linked with a DNA linker. The larger nanoparticle (A) accommodates many small nanoparticles (B) around it, producing a so-called ‘satellite’ ABn system. Interconnections between these ‘satellite’ aggregates yield a 3D-network (ABn)m. An important factor controlling the network optical properties is the length of the DNA oligonucleotide: It controls not only the inter particle distances, but also the rate of aggregation and thus the number of nanoparticle per aggregate.
In a similar approach, DNA-functionalized CdSe/ZnS nanoparticles and Au nanoparticles were cross-linked with a complementary linker to yield a binary network. Fluorescence and absorption spectroscopy indicated cooperative optical and electronic phenomena within the resulting material. Changes in the optical and electronic properties of nanoparticles upon their aggregation provide a way to detect the association process, thus allowing DNA- biosensor construction.
The application of DNA-functionalized Au Nano rods instead of spherical nanoparticles results in micro sized rods composed of many cross-linked Nano-units. This material offers unusual anisotropic properties. Long DNA molecules capable of accommodating several complementary DNA fragments in different domains on the strand can serve as templates for the construction of specific linear assemblies. In order to prevent the microscopic crosslinking of nanoparticles resulting in a network, Au nanoparticles functionalized with a single maleimide group were reacted with thiolderivatized DNA oligonucleotides.
The resulting mono- DNA-functionalized Au nanoparticles were associated with the respective complementary domains on the DNA template molecule. Individual placement of the nanoparticles at specific complementary domains of the template allows control of distances between them, as well as controlled placement of different kinds of nanoparticles (different sizes, different materials) on the template, which results in a variety of properties of the nanoparticles assemblies. For example, two DNA functionalized Au nanoparticles can be placed on the DNA template with three different orientations: ‘head-to-head’, ‘head-to- tail’, and ‘tail-to-tail’, thus providing different distances between the nanoparticles.
Composite Assemblies of Nucleic Acids, Proteins, and Nanoparticles:
A combination of the synthetic methods to prepare biomaterial-functionalized nanoparticles and the application of proteins with affinity properties (e.g. Sav, antibodies), oligonucleotides and nanoparticles as building blocks, allows the construction of very sophisticated bio-molecular/nanoparticle clusters.
In order to functionalize Au nanoparticles with antibodies, a multi-step procedure was performed. 50-Thiolmodified oligonucleotide was covalently bound to Sav using the hetero-bio-functional cross-linker sulfosuccinimidyl-4- (maleimidomethyl) cyclohexane-1 -carboxylate.
The resulting conjugate has four binding sites for biotin ligands and one nucleic acid function. The Sav receptor sites were reacted with biotinylated antibodies directed against mouse IgG (aM-IgG) or rabbit IgG (aR-IgG), resulting in DNA-tagged antibody conjugates that can bind to the complementary DNA from one side and to the respective antigens from the other. This architecture was reacted with Au nanoparticles functionalized with oligonucleotides complementary to the DNA tag, thus finally resulting in the Au nanoparticles modified with the antibody molecules.
The resulting conjugate was then applied as a label for the immunoassay of the respective antigens. The linkage of the Au nanoparticle to the antigen-antibody complex was further amplified by the electro less deposition of Ag on the Au nanoparticles. A combination of receptor proteins and oligonucleotides as binding blocks for the directed aggregation of nanoparticles allows the complex structuring of the assemblies. For example, Sav was interacted with biotinylated DNA, resulting in a four DNA-Sav conjugate.
Application of this conjugate as a building block in the aggregation of DNA-functionalized Au nanoparticles yields a network with a motif different from that resulting from direct aggregation of DNA-functionalized nanoparticles bound by the complementary oligonucleotide.
Even more fascinating aggregation was achieved when Au nanoparticles were primarily associated with Sav to yield Nano-clusters that were then placed on a DNA template. In this work, 1.4 nm Au nanoparticles with a single amino substituent were derivatized with a biotin group, and the biotin moiety was used to organize the Nano-clusters into the tetrahedral superstructure defined by the geometry of the biotin-binding sites of the Sav.
Prior to cluster formation, Sav was derivatized with a single DNA molecule. The resulting tetrahedral four-Au nanoparticle clusters, each with a single DNA tail, were assembled on a DNA template molecule with positions and distances dictated by the sequence of the DNA template. This approach could allow the formation of almost any imaginable structure composed of various nanoparticles of different sizes and materials.
An interesting application of submicron metallic ‘barcodes’ for the analysis of biomaterials such as DNA or antigens has recently been reported. ‘Barcodes’ a few hundred nanometers across and several microns long composed of many different metals (Pt, Pd, Ni, Co, Ag, Cu, and Au) were prepared by the electrochemical deposition of the metals in a-porous Al2O3 membrane (Fig. 9.5).
The structure of the barcode could be identified by optical reflectivity or field emission-SEM. The use of a mixture of ‘barcodes’, each used to identify a specific biomaterial adsorbed on it, could enable the simultaneous analysis of numerous components in biological mixtures. Thus, the barcode concept adds a nanoparticle based analysis method that competes with chip-arrays and encoded micro-beads. In addition, as the tag identification can be performed optically, fluorescence can be used to characterize a bio-recognition event.
Assembly of Biomaterial-Nanoparticle Architectures on Surfaces:
The immobilization of biomaterials on transducers such as electrodes, piezoelectric crystals, and field-effect transistors attracts research efforts directed to the development of biosensors and bio-electronic devices. Several methods for the surface functionalization of electronic elements have been developed, and means to couple the active sites or recognition processes of biomaterials with the electronic elements, giving electronic transduction of the biological events, have been demonstrated.
Biosensors, biofuel cell elements and opto-bio-electronic systems have been constructed. Similarly, the deposition of biomaterials on non-conductive surfaces (e.g. plastics, glass), and especially the pattering of such surfaces with biomaterials, is of high interest in tailoring protein-chips and DNA-chips for proteomic or genomic analyses.
Also, diverse methods for the assembly of 2D and 3D nanoparticle structures on surfaces have been developed. Various applications of these systems in the fabrication of photo- electrochemical cells (e.g. solar cells), light- emitting diodes, electro-chromic systems, and computing devices, and also the organization of different Nano-metric single-electron devices, have been discussed.
The integration of nanoparticle-biomaterial hybrid systems with surfaces paves the way for the generation of ordered architectures with new functionalities. The unique photonic properties of nanoparticles may be employed to detect and probe biological recognition events on surfaces.
The spectroscopic properties of molecular adsorbents on nanoparticle surfaces such as surface Plasmon resonance (SPR), surface- enhanced Raman spectroscopy (SERS), and surface-enhanced fluorescence may be used to probe biomaterial structures on surfaces. Also, the unique electronic (e.g., conductive) and catalytic properties of nanoparticles may be used to provide electron transfer from or to redox-centres of enzymes, or to enhance chemical re-activities of or on the biomaterial structures.
From another direction, biomaterial constituents on surfaces provide unique features for the formation of the hybrid biomaterial- nanoparticle structures. Biomaterials (e.g. nucleic acids, streptavidin-biotin antigen- antibody complexes) assembled on surfaces can provide templates for the immobilization of nanoparticles.
Enzymes may be used as catalytic tools for the manipulation of biomaterials on surfaces. For example, the ligation, scission or polymerization of nucleic acids may be used to tailor nucleic acids of specific lengths and compositions. Thus, by the genetic manipulation of biomaterials, the synthesis of nucleic acids, and the bio-catalytic manipulation of the biomaterials on surfaces, complex biomaterial-based structures on surfaces may be engineered.
The incorporation and integration of nanoparticles in biological templates can then yield new functional materials. For example, the catalytic deposition of metals on nanoparticle seeds can generate circuitry with the shape of the biomaterial.
Thus, hybrid biomaterial nanoparticle composites on surfaces provide functional interfaces that could be important materials for bio-electronic, electronic, optobioelectronic and photonic applications. The following sections address the methods of organizing these composite systems on surfaces and discuss recent activities that utilize these systems for sensoric, photo electrochemical and electronic circuitry applications.
Assembly of Layered Nanoparticle-Protein Assemblies:
Multilayer systems assembled on solid supports and composed of cytochrome c (Cytc) molecules and Ti02 nanoparticles layers have been fabricated using the layer-by-layer deposition of the biological and inorganic components. The TiO2 nanoparticle layers were deposited from aqueous solution by boric acid-promoted hydrolysis of hexafluorotitanate ion (liquid phase deposition process, LPD) (Fig. 9.6).
The LPD technique enables the preparation of anatase TiO2 without employing organic solvents. This is essential for co-deposition of the biomaterial. The TiO2 nanoparticles produced by this technique on the surface are negatively charged. The subsequent deposition of the positively charged protein cytochrome c (isoelectric point, pI = 10.1) results in electrostatic attraction between the TiO2 nanoparticles and the protein, allowing multilayer formation.
The deposition of the multilayers was followed by quartz crystal microgravimetry (QCM), demonstrating a linear increase in the mass upon deposition of each double layer TiO2/Cyt c, which was verified by absorbance spectroscopy. This method could be widely extended to the preparation of other protein/metal oxide multilayers that would facilitate the creation of photo-functional protein/inorganic super lattices.
Nucleic Acid-Nanoparticle Architectures on Surfaces:
Nucleic acids can serve as templates capable of binding DNA-functionalized nanoparticles at complementary segments. When DNA templates are fixed at a surface of a solid support, the resulting assemblies of nanoparticles can yield a pattern dependent either on the shape produced by the DNA template itself or on the pattern produced upon its immobilization.
A natural single-stranded long-chain virus-DNA (M13mp18, 7246 bases) was hybridized with a biotinylated short-chain target oligonucleotide complementary to a specific segment of the viral DNA. It should be noted that the template viral DNA can have more than one complementary domain, and thus is capable of accommodating more than one biotinylated-DNA chain.
The hybridized DNA-holding biotin units were then reacted with Sav-coated Au nanoparticles (5 nm). The resulting negatively charged Au nanoparticle- labeled DNA template was adsorbed on a surface of positively charged Mg2+-coated mica substrate and was subjected to atomic force microscopy (AFM).
The images obtained clearly showed that Au nanoparticles associated with the DNA template, yet the number of Au nanoparticles per DNA chain was not perfectly defined, reflecting either incomplete hybridization of the template with the biotinylated target DNA or incomplete association of the Sav-functionalized Au with the biotinylated domains.
As different biotinylated nucleic acid segments may be hybridized with the M13mp18DNA, the entire viral DNA may be decorated with Sav-Au nanoparticles. Indeed, such an experiment revealed that the Au nanoparticle-functionalized viral DNA was formed on the solid support.
The electrostatic association of CdS nanoparticles with DNA was applied as the driving force for the formation of the semiconductor nanoparticle DNA wire on the surface. Negatively charged ds-DNA molecules were located at the water-air interface containing cationic surfactant molecules.
The layer at the interface was compressed using the Langmuir-Blodgett technique in order to provide a high-density packing of the DNA molecules. Negatively charged CdS nanoparticles (3 nm) capped with thiocholine were then added to the aqueous phase, resulting in their aggregation with the DNA.
The obtained DNA/CdS aggregates were transferred to a solid support, and electron microscopy images revealed the formation of densely packed CdS Nano-parties along the DNA template. These generated chains are composed of semi-conductive nanoparticles with a diameter of ca. 3 nm, the average distance between the centres of the adjacent nanoparticles being ca. 3.5 nm.
The circular plasmid ds-DNA pUCLeu4 (3.455 base pairs) was reacted with Cd2+ ions, and the resulting Cd2+/DNA complex was adsorbed onto a polylysine-coated glass slide by spin coating. The complexes Cd2+ ions were reacted with S2- to yield CdS, and electron microscopy images revealed the formation of CdS nanoparticles that followed the circular shape of the DNA template. In another example, positively charged lysine-capped Au nanoparticles were deposited onto a surface coated with negatively charged thick DNA film.
Electrostatic interaction between the positive charges on the Au nanoparticles and the negative charges on the phosphate groups of the DNA template molecules leads to the assembly of nanoparticles in linear superstructures. The DNA molecules, which are locked into a fairly rigid structure prior to addition of the lysine-capped Au nanoparticles, are rendered mobile by solvation during the addition of the nanoparticle solution to the DNA film surface. This process facilitates reorganization into highly regular linear nanoparticle superstructures during electrostatic complexation.
AFM and STM visualization of surface-confined DNA molecules can be greatly improved by the addition of complementary DNA-functionalized Au nanoparticles that not only introduce clearly visible Au clusters associated with the DNA lying on the surface but can also change the orientation of the long DNA molecules at the surface, producing tethered rather than flat-lying DNA chains. Both these effects can facilitate the visualization of DNA molecules at surfaces.
Negatively charged DNA molecules can complex positively charged metal ions that are later reduced to the metallic state, producing a metal wire across the DNA template. For example, this was accomplished by loading the DNA template with Pd2+ cations and adding reductive chemicals afterwards, leading to the formation of Nano-scale Pd clusters on the DNA.
Depending on the duration of the reduction process, well-separated Pd clusters or a quasi-continuous Pd wire with a grain-like structure could be produced. A system utilizing DNA that crosslinks into a continuous network has been investigated in an attempt to improve structural control over a longer distance (Fig. 9.9).
Three types of oligonucleotides were used, labeled 36, 37 and 38. Oligonucleotide 36 had a sequence of 17 bases, and its 3′ end was modified with thiol group (5′-GTAAAACGACGACGGCCA- GT-SH-3′). A 57-base oligonucleotide (37) included a central sequence complementary to 36 and sides of alternating A and T bases. Oligonucleotide 38 was 50 base poly (dAdT). This oligonucleotide forms a network structure and is hybridized upon contact with 37.
All three oligonucleotides were mixed with Au nanoparticles (5 nm). The resulting network was adsorbed onto a mica surface, and AFM images were obtained. These images resembled a Nano circuit, but it lacked organization and direct contact between the Au nanoparticles.
In order to construct real micro-sized DNA- templated wires, micro-wires prepared by electrochemical metal deposition in a porous alumina membrane were bound to thiolated DNA molecules and reacted with a solid support functionalized with a complementary DNA oligonucleotide. The hybridization process between the surface-confined DNA and the DNA linked to the micro wires yielded ds- DNA bridges binding the micro wires to the surface.
It should be noted that in this work the DNA molecules were used as ‘molecular glue’ to bind micro-sized predesigned wires to the surface instead of individual nanoparticles, thus providing a significant advance in micro-scale circuitry.
Another approach to the directed placement of nanoparticles on solid supports by means of DNA is based on the micro patterning of a surface with DNA molecules. This approach provides the specific binding of complementary DNA-functionalized nanoparticles at specific domains of the pattern. An amino derivatized oligonucleotide was deposited onto a chemically modified glass surface in a form of a pattern by a Nano liter dispensing device, and was then covalently bound to the surface.
Au nanoparticles (34 nm) functionalized with a complementary oligonucleotide were then reacted with the DNA-patterned surface, yielding an Au nanoparticle pattern that follows the shape of the DNA pattern. Even more precise placement of Au nanoparticles was achieved by the ‘dip-pen nanolithography’ that was applied for patterning the primary DNA on the surface. Two kinds of amino-functionalized DNA oligonucleotides, TCTCAACTCGTAA10 (39) and A10CGcat- tcaggat (40), were deposited onto a 1,16-mercaptohex-adecanoic acid-functionalized Au surface, and were then covalently bound to it (Fig. 9.10).
The deposition was performed in two sequential steps to yield a pattern composed of spots of both oligonucleotides. This primary pattern was reacted with an oligonucleotide of sequence TACGAGTTgAGaatcct- gaatgCG (41) composed of two domains: one complementary to 39 and the other to 40. The resulting DNA-functionalized Surface provided complementary DNA chains for two kinds of DNA-functionalized Au nanoparticles: 39 Au (13 nm) and 40 Au (31 nm).
A pattern was obtained on the DNA-functionalized surface: 39 Au nanoparticles specifically bound to the spots of 40/41 and 40 Au nanoparticles bound to the spots of 39/41 periodically located on the solid support. Multilayers of nanoparticles can be assembled on solid supports by utilizing DNA complementarity. For this purpose, a glass surface was functionalized with a monolayer of an oligonucleotide (42), and then the surface was reacted with an oligonucleotide (43) composed of two domains, one of which was complementary to 42 while the other provided complementarity for another oligonucleotide (44).
Oligonucleotide 44-functionalized Au nanoparticles (13 nm) were added to yield a monolayer of ds-DNA (ds-43/44) attached to Au nanoparticles (Figure 9.11). Since many of the DNA chains (44) around the Au nanoparticle were not reacted with the surface-confined DNA (43), they could be reacted with further complementary domains of oligonucleotide 43, thus providing binding sites for 42.
A second kind of Au nanoparticle modified with oligonucleotide 42 was reacted with this first layer assembly, resulting in a second layer of Au nanoparticles. The repetitive stepwise deposition of Au nanoparticles functionalized with the oligonucleotides 42 and 44 resulted in a multilayer assembly of Au nanoparticles.
Each hybridized Au nanoparticle layer imparted a deeper red colour onto the substrate. It should be noted that this method could allow layer-by-layer deposition of different (by size or by chemical nature, e.g., Ag, CdS, CdSe) kinds of nanoparticles.
Functional Biomaterial-Nanoparticle Structures on Surfaces for Sensory and Electronic Applications:
The unique properties of Nano-partides make Nano-partides-biomaterial conjugates attractive labels for sensing applications. The optical and electronic sensing of biomaterials on surfaces is a common practice in analytical biochemistry. Thus, the immobilization of Nano-partides-biomaterial conjugates on surfaces provides a general route for the development of optical or electronic biosensors. Also, the immobilization of metallic or semiconductor Nano-partides-biomaterial conjugates gives a method for the assembly of electronic circuitry.
In such a system, the biomaterial could provide a structural template for the electronic circuitry. The photonic and optical properties of the nanoparticles enables imaging of the structural features of the circuitry, whereas the catalytic functions of the nanoparticles allow their metallic enlargement to become interconnected with long-range conductivity.
Nanoparticle-Biomaterial Conjugates for Optical Sensing and Analysis:
Au nanoparticles linked to bio-receptors provide labeled conjugates that can be used to follow the bio recognition event at a biosensor surface. Various optical methods have been employed to detect the association of Au nanoparticles to biochip domains, including, for example the scanometric detection of light scattering, surface Plasmon resonance, resonance-enhanced absorption by nanoparticles, nanoparticle fluorescence, and enhanced Raman scattering.
Although not limited to DNA detection, most of these detection schemes were applied for this purpose. In a typical setup for scanometric detection, a modified glass slide was mounted on a microscope stage and illuminated in the plane of the slide with white light.
In this configuration, the slide served as a planar waveguide, preventing any light from reaching the microscope objective by total internal reflection. Wherever nanoparticle probes were attached to the surface, however, evanescently coupled light was scattered out of the slide and was seen as brightly coloured spots. This approach was used for the detection of Au nanoparticle-labeled DNA molecules specifically bound to a DNA-functionalized surface.
Enlargement of the Au nanoparticles by the reduction of silver salts on them allows a hundred-fold amplification of the signal, and thus increases the sensitivity. This method was used to detect single-base mismatched DNA oligonucleotides immobilized at different domains of the glass support. The high sensitivity required was provided by the silver deposition, and selectivity was achieved by examining the ‘melting’ profiles of the spots (the mismatched spot has a lower ‘melt’ temperature due to its lower association constant).
Two different DNA sequences could be detected at once when two Au nanoparticles of different sizes were used to label different oligonucleotides. Spots with different colours corresponding to two kinds of Au nanoparticles were detected on the glass support when domains functionalized with different DNA samples were reacted with the corresponding complementary DNA labeled with Au nanoparticles 50 nm and 100 nm in diameter.
Ag nanoparticles have similarly been used as antibody labels for the detection of antigens. This setup provided sensitivity high enough to perform immunoassays based on single-target detection. Resonance enhancement of the absorptive properties of metal nanoparticles bound to a surface by bio-recognition interactions was also used as an effective means for biosensor devices.
Au nanoparticles have been used extensively for the detection of bio-affinity binding by virtue of enhanced surface Plasmon resonance (SPR). A dramatic enhancement of SPR bio-sensing was achieved for immunosensing and DNA sensing when Au nanoparticles were used.
A sensor interface was modified with antibody units, the surface was reacted with the complementary antigen component, and finally this affinity assembly was reacted with a secondary antibody labeled with an Au nanoparticle. The association of antigen with an antibody-functionalized surface can be detected by SPR. However, the change in the SPR spectrum is larger if the secondary antibody is bound to the surface.
It should be noted that the secondary antibody was able to associate with the modified surface only when the antigen was already bound there, providing an amplification route for the primary recognition event. Amplification is dramatically increased when the secondary antibody is labeled with an Au nanoparticle.
The binding of the Au nanoparticle to the immunosensing interface led to a large shift in Plasmon angle, a broadened Plasmon resonance, and an increase in the minimum reflectance, thereby allowing pico-molar detection of the antigen.
Similarly, an enhancement in sensitivity by 3 orders of magnitude was obtained in DNA analysis when Au nanoparticle-functionalized DNA molecules were used as probes. Surface-enhanced Raman scattering (SERS) of nanoparticle bound substrates allows the amplification of molecular vibrational spectra up to 105 fold.
Although the technique has not been widely applied for detecting bio-recognition events on surfaces, it was demonstrated that a Cytochrome c-Au nanoparticle conjugate associated with an Ag surface revealed an SERS spectrum. Semi-conductive nanoparticles (e.g., CdS, CdSe) can be used as fluorescent labels for immunosensing and DNA sensing, providing tunable wavelength, narrow emission peaks, and hundred-fold higher stability than molecular fluorescent dyes.
Electronic Transduction of Bio-Sensing Events by Nanoparticle-Biomaterial-Functionalized Arrays:
Redox-enzymes can be contacted with electrodes by the tethering of redox units to the proteins or by the surface reconstitution of apo-proteins on electron-relay cofactor units. Recently, the electrical contacting of redox-enzymes was accomplished by the reconstitution of the apo-flavoenzyme apo-glucose oxidase (apo-GOx) with a 1.4 nm Au-nanoparticle functionalized with N6- (2-aminoethyl)-flavin adenine dinucleotide (FAD cofactor amino-derivative).
The conjugate produced was assembled on a thiolated monolayer using various dithiols as linkers. Alternatively, the FAD-functionalized Au nanoparticle could be assembled on a thiolated monolayer associated with an electrode, with apo-GOx subsequently reconstituted on the functional nanoparticle. The enzyme-electrodes prepared by these two routes reveal similar protein surface coverage of 1 x 10-12 mole cm-2.
The nanoparticle-reconstituted glucose oxidase layer is electrically contacted with the electrode without any additional mediators, and it stimulates the bio- electro-catalysed oxidation of glucose. The resulting nanoparticle-reconstituted enzyme electrodes reveal unprecedentedly efficient communication with the electrode (electron transfer turnover rate 5000 s-1) this electrical contacting makes the enzyme-electrode insensitive to oxygen or to common oxidizable interferon’s such as ascorbic acid.
A related electrochemical method was employed for the Au nanoparticle-based quantitative detection of the 406-base human cytomegalovirus DNA sequence (HCMV DNA). The HCMV DNA was immobilized on a micro well surface and hybridized with an oligonucleotide-modified Au nanoparticle.
The resulting surface-immobilized Au nanoparticle double-stranded assembly was treated with HBr/Br2, resulting in the oxidative dissolution of the gold particles. The solubilized Au3+ ions were electrochemically reduced and accumulated, and then determined by anodic stripping voltammetry using a sandwich-type screen-printed micro band electrode (SPMBE).
The combination of the sensitive detection of Au3+ ions at the SPMBE by non-linear mass transfer of the ions and the release of a large number of Au3+ ions upon the dissolution of the particle associated with a single recognition event provides an amplification path that enables the detection of the HCMV DNA at a concentration of 5×10-12 M.
The conductivity stimulated by patterned nanoparticle bridges provides an alternative route for designing miniaturized biosensors. This was exemplified by the design of a miniaturized immuno-sensor based on Au nanoparticles and their catalytic properties (Fig. 9.12).
Latex particles stabilized by an anionic protective layer are attracted to a gap between micron-sized Au electrodes by the application of a non-uniform alternating electric field between the electrodes (di-electrophoresis). Removal of the protective layer from the latex particles by an oppositely charged poly-electrolyte results in the aggregation of the latex particles and their fixation in the gap.
Adsorption of protein A on the latex surface yields a sensing interface for the human immunoglobulin (IgG) antigen. The association of human immunoglobulin on the surface is probed by the association of the secondary Au-labeled anti-human IgG antibodies to the surface, followed by the catalytic deposition of a silver layer on the Au particles.
The silver layer bridges the gap between two microelectrodes, resulting in a conductive ‘wire’. Typical resistances between the microelectrodes were 50-70Ω whereas control experiments generated resistance > 103Ω. The method enabled the analysis of human IgG with a detection limit of 2 x 1013M.
DNA detection has also been performed using microelectrodes fabricated on a silicon chip. A probe nucleic acid was immobilized on the SiO2 interface in the gap separating the microelectrodes. The target 27-mer nucleotide was then hybridized with the probe interface, and subsequently a nucleic acid-functionalized Au nanoparticle was hybridized with the free 3′-end of the target DNA.
The Au nanoparticle catalysed hydroquinone-mediated reduction of Ag+ ions, resulting in the deposition of silver on the particles, lowering the resistance between the electrodes. Single-base mutants of the analyte oligonucleotide were washed off from the capture-nucleic acid by the application of a buffer of an appropriate ionic strength, so were not detected.
A difference of 106 in the gap resistance was found between the analyte and the mutants. The low resistances between the microelectrodes were found to be controlled by the concentration of the target DNA, and the detection limit for analysis is in the region of 5 x 10-13 M. This sensitivity translates to 1 mg. ml/1 of human genomic DNA or ca. 0.3 mg. mL-1of DNA from a small bacterium. These concentrations suggest that the DNA may be analysed with no pre-PCR amplification.
Photo electrochemical transduction of DNA recognition processes has been shown by use of semiconductor (CdS) nanoparticles modified with nucleic acids. Semiconductor CdS nanoparticles (2.6 ± 0.4 nm) were functionalized with the two thiolated nucleic acids that are complementary to the 5′ and 3′ ends of a target DNA.
An array of CdS nanoparticle layers was constructed on an Au electrode by a layer-by-layer hybridization process. Illumination of the array resulted in the generation of a photocurrent. The photocurrent increases with the number of CdS nanoparticle layers, and the photocurrent action spectra follow the absorbance features of the CdD nanoparticles, implying that the photocurrents originate from the photo-excitation of the CdS nanoparticles.
That is, photo-excitation of the semiconductor causes the formation of an electron-hole pair. Transfer of conduction donor to the valence band holes, yields a steady-state photocurrent in the system. The ejection of the conduction-band electrons into the electrode occurred from nanoparticles in intimate contact with the electrode support.
This is supported by the fact that Ru(NH3)6+ units (E = -0.16 V vs. SCE) that are electrostatically bound to the DNA enhance the photocurrent from the DNA-CdS array. That is, the Ru(NH3)63+ units act as an electron wiring element that facilitates electron hopping to the electrode through the DNA tether from CdS particles that lack contact with the electrode.
The system is important not only as it demonstrates the use of photo electrochemistry as a transduction method for DNA sensing, but also since the system reveals the Nano-engineering of organized DNA-tethered semiconductor nanoparticles on conductive supports. These latter Nano engineered structures are the first step toward electronic Nano circuitry.
The immobilization of nanoparticles on surfaces may also be used to yield high surface area electrodes. Enhanced electrochemical detection of nucleic acids was reported by the roughening of flat gold electrodes with an Au nanoparticle monolayer. The roughening of an Au-quartz crystal with a monolayer consisting of Au nanoparticles has also been employed for the enhanced micro gravimetric analysis of DNA.
Biomaterial-Nanoparticle Arrays for Electronic Circuitry:
DNA molecules can be loaded with metallic or semi conductive nanoparticles, which form wires or networks upon their deposition onto a solid. In order to use these nanowires for electronic circuitry, they should be electrically connected to the external world, and they need to be conductive.
Since nanoparticles are loaded onto the DNA template with gaps between them, the issue of electrical conductivity is important. Electron transport through DNA has been one of the most intensively debated subjects in chemistry in the first decade of this century, and it is still under extensive theoretical and experimental investigation.
In order to examine the electrical conductivity of ds-DNA spacers connecting Au nanoparticles, thiol-derivatized oligonucleotides were adsorbed on Au nanoparticles, and then the DNA-functionalized Au nanoparticles were bridged with DNA chains composed of double-stranded helices of various lengths.
These helices were terminated on both sides with single-stranded domains complementary to the oligonucleotides bound to the Au nanoparticles. The resulting Au nanoparticle aggregates linked with ds-DNA spacers were deposited on an electrically non-conductive solid support, and their conductivity was measured by the four-probe method.
Surprisingly, the conductivities of the aggregates formed from all three linkers ranged from 10-5 to 10-4 S cm-1 at room temperature, and they showed similar temperature-dependent behaviour. The similarity of the electrical properties of the aggregates originates from the fact that the DNA spacers are compressed, thus providing small and similar distances between the Au nanoparticles.
Accordingly, the measured conductivity parameters reflect the electrical properties of the metallic nanoparticles separated by short gaps. The conductivity of metallic nanoparticle aggregates can be enhanced upon the chemical deposition of another metal (e.g., Ag deposition on Au aggregates).
DNA-functionalized Au nanoparticles were placed between microelectrodes by hybridization with complementary DNA molecules immobilized in the non-conductive gap between two electrodes. Then the nanoparticles were subjected to the reductive deposition of Ag to enlarge the metallic species and bridge the gaps between them with deposited Ag.
The subsequent value of conductivity was found to be dependent on the original loading of the gap with Au nanoparticles, thus allowing the quantitative measurements of the DNA-functionalized Au nanoparticles bound to the surface.
A single nanowire produced on a DNA template bridging two micro-size electrodes was constructed. Two micro-electrodes facing each other (12-16 mm separation) were functionalized with 12 base oligonucleotides that were then bridged with a 16 mm long 1-DNA (Fig. 9.13).
The resulting phosphate units of the DNA bridge were loaded with Ag+ ions by ion-exchange, which were then reduced to Ag metal by hydroquinone. The small Ag aggregates produced along the DNA backbone were then used as catalysts for further reductive deposition of silver, eventually leading to the formation of an Ag nanowire. This micrometer-sized element had a typical width of 100 nm and a granular morphology (as determined by AFM).
Electrical measurements revealed non-linear I-V curves. A similar approach was used to generate highly conductive nanowires bridging macroscopic Au electrodes (198). The DNA molecules were positioned between macroscopic Au electrodes and loaded with Pd2+ ions. The ions were then chemically reduced to continuous metallic wires, providing specific conductivity only one order of magnitude below than of bulk palladium.
Biomaterial-Functionalized Magnetic Particles:
Magnetic particles (microspheres, Nano-spheres, and Ferro fluids) are widely studied and applied in various fields of biology and medicine such as magnetic targeting (drugs, genes, radiopharmaceuticals), magnetic resonance imaging, diagnostics, immunoassays, RNA and DNA purification, and cell separation and purification.
These magnetic particles are generally of the ‘core-shell’ type: biological species (cells, nucleic acids, proteins) are connected to the magnetic ‘core’ through organic linkers, often organized as a polymeric ‘shell’ around the core. The magnetic core is usually composed of Fe3O4, and its primary modification with an organic ‘shell’ can include the adsorption of an organic polymer or the covalent attachment of organosilane molecules.
Organic functional groups introduced by these techniques allow the coupling of biomolecules to the organic ‘shell’. Several synthetic approaches have been applied to couple artificial magnetic particles to biomolecules, which were then used for various bio- analytical applications. For example, antibody molecules were adsorbed on synthetic Fe3O4 magnetic particles and were then used for specific binding to cells and the separation of the cells in an external magnetic field.
DNA molecules were reacted with a mixture of Fe2+ and Fe3+ ions that were electrostatically associated with DNA chains. These iron ions were then chemically reacted to give Fe3O4 magnetic particles associated with the DNA molecules. The labeled DNA could hybridize with complementary oligonucleotides, and the magnetic particles linked to the DNA molecules allowed the separation of the labeled from the non-labeled DNA strands.
The natural protein ferritin can be used to generate precisely sized and shaped magnetic particles in a protein cavity. For this purpose, the non-magnetic natural core composed of 5Fe2O3, 9H2O was chemically removed from the protein cavity, and an artificial magnetic core composed of magnetite (Fe3O4) or magnetite/maghemite (Fe3O4/g-Fe2O3) was generated inside it.
The natural protein shell remains, providing binding groups for the covalent attachment of biomolecules. Some bacteria (e.g. Magnetospirillum magnetotacticum MS-1, Magnetospirillum sp. MGT-1, Magnetospirillum sp. AMB-1, and Magnetospirillum gryphiswaldense) produce natural magnetic particles (Fe3O4, 50-100 mm in size) aligned in chains and enveloped by a lipid membrane.
ADVERTISEMENTS:
These naturally produced lipid covered magnetic particles can be isolated from the parent bacteria and modified with biomolecules using bio-functional coupling reagents (e.g., glutaric dialdehyde). They have been applied for fluoroimmunoassay, chemiluminescence immunoassay, mRNA recovery, and as DNA carriers.
Most of these techniques are based on the magnetic separation of the magnetic nanoparticle-linked biomolecules from the non-labeled biomolecules. The immobilization of enzymes on magnetic particles yields bio-catalytically active particles. In one example, alcohol dehydrogenase was covalently bound to Fe3O4 magnetic particles, and the immobilized enzyme continued to demonstrate high bio-catalytic activity.
Most of the applications of biomaterial-magnetic particle hybrid conjugates involve the concentration, separation, regeneration, mechanical translocation, and targeting of biomaterials. As these subjects do not deal directly with nanoparticle biomaterial conjugates, we will not cover them here.
Some research being carried on now (2012), however, has been directed to the coupling of functional magnetic particles with biomaterial to yield magnetically-controlled functions. Magnetic particles were modified with redox-relay groups such as ferrocene or bi-pyridinium units and applied for the magneto-switched activation or deactivation of the bio-electro-catalytic functions of redox-enzymes. Ferrocene functionalized magnetite particles were used for the magnetic switching of the bio-electro-catalysed oxidation of glucose in the presence of glucose oxidase (GOx).
Magnetic attraction of the particles to the electrode enabled the oxidation of the ferrocene-linked redox units that mediated the GOx-bio-catalysed oxidation of glucose. Positioning of an external-magnet above the electrochemical cells lifts the magnetic particles from the electrode, and the bio-electro-catalysed oxidation of glucose is switched off.
Similar magnetic switching of the bio-electro-catalysed reduction of nitrate in the presence of nitrate reducatase (NR) was accomplished by the application of N,N’-bipyridinium-functionalized magnetic particles.
Similarly, magnetic nanoparticles linked to nicotinamide adenine dinucleotide through PQQ were used for the magnetic switching of the bio-electro-catalysed oxidation of lactate to pyruvate using lactate dehydrogenase (LDH).
These particles were employed for the dual selective of lactate and glucose in the presence of LDH and GOx. An Au electrode was functionalized with a ferrocene-labeled monolayer, and PQQ- NAD+ modified particles were suspended in electrolyte solution. Removal of the magnetic particles from the electrode by means of the external magnet enables the electrochemical detection of glucose at potentials more positive than 0.32 V (vs SCE) by the ferrocene-mediated bio-electro-catalyzed oxidation of glucose in the presence of GOx. Attraction of the functional magnetite particles to the electrodes by the external magnet enables the amperometric analysis of lactate in the potential range -0.13 V < E < 0.32 V.
A novel approach to DNA analysis involved the use of magnetic particles modified by a nucleic acid that hybridizes with a bio-tinylated target DNA. The resulting hybridized complex was then reacted with a streptovid-in-Au nanoparticle conjugate.
A silver shell was catalytically generated on the Au nanoparticles linked to the DNA molecules, and then the magnetically labeled assembly was moved to an electrode by means of an external magnet, where electrochemical stripping of the Ag layer provided an amperometric signal for the quantitative determination of the analyte DNA.
The mechanical rotation of magnetic particles using an external rotating magnet has also been used as an amplification method for electrochemical sensing. In these systems, chemical transformations are controlled by convection of the solution and its constituents rather than by diffusion.
As a result, mass transport to the magnetic particles is controlled by the rotational speed of the particles around the electrode axis. Pyrroloquinoline Quinone functionalized magnetite particles were employed for the electro-catalysed oxidation of NADH under rotation.
The electro-catalytic anodic currents are controlled by the circular rotation speed of the particles. Similarly, the ferrocene functionalized particles were used to control the bio-electro-catalysed oxidation of glucose in the presence of GOx by means of the circular rotation of the particles produced by the external magnetic rotor;the bio-electro-catalytic current provided by the soluble GOx and ferrocene mediator linked to the rotating magnetic particles were dependent on the particle rotation speed. The calibration plots derived from the cyclic voltammograms show a dramatic influence of the particle rotation speed on the sensitivity of the bio-electro-catalytic system to glucose.
This method represents a novel route to the amplification of bio-electronic detection schemes or the enhancement of biological recognition processes. The labelling of biological components by magnetic particles (e.g., antibodies or nucleic acids) and their external rotation by means of the magnetic rotor may enhance the formation of antigen-antibody complexes or hybridization, respectively, by the agitation of the solution.
Similarly, bio-recognition events transduced by means of bio-electro-catalytic labels may be amplified by coupling the bio-electro-catalytic label to magnetic labels and rotating the conjugated bio-recognition complexes with an external magnetic rotor.