ADVERTISEMENTS:
This article throws light upon the top three cutting-edge techniques used in molecular biology.
The top three cutting-edge techniques are: (1) Fluorescent In Situ Hybridization (FISH) (2) Blotting Techniques and (3) Polymerase Chain Reaction.
Technique # 1. Fluorescent In Situ Hybridization (FISH):
FISH (Fluorescent in situ hybridization) is a cytogenetic technique which can be used to detect and localize the presence or absence of specific DNA sequences on chromosomes. It uses fluorescent probes which bind only to those parts of the chromosome with which they show a high degree of sequence similarity.
ADVERTISEMENTS:
FISH has evolved into a core technique spanning many areas of diagnosis and research, including cancer cytogenetics, prenatal screening of chromosomal aberrations, molecular pathology and developmental molecular biology.
The general principle of FISH, hybridizing fluorescently-labeled loci-specific, telomeric or centromeric DNA probes to chromosomes, can be applied to a range of specimens such as metaphase chromosome spreads or to inter-phase nuclei and cells.
Fluorescence microscopy can be used to find out where the fluorescent probe bound to the chromosome. FISH is often used for finding specific features in DNA. These features can be used in genetic counseling, medicine, and species identification.
First, a probe is constructed. The probe has to be long enough to hybridize specifically to its target (and not to similar sequences in the genome), but not too large to impede the hybridization process, and it should be tagged directly with fluorophores, with targets for antibodies or with biotin. This can be done in various ways, for example nick translation and PCR using tagged nucleotides.
ADVERTISEMENTS:
Then, an inter-phase or metaphase chromosome preparation is produced. The chromosomes are firmly attached to a substrate, usually glass slides. Repetitive DNA sequences must be blocked by adding short fragments of DNA to the sample.
The probe is then applied to the chromosome DNA and incubated for ~12 hours while hybridizing. Several wash steps remove all un-hybridized or partially hybridized probes. The results are then visualized and quantified using a microscope that is capable of exciting the dye and recording images.
If the fluorescent signal is weak, amplification of the signal may be necessary in order to exceed the detection threshold of the microscope. The signal strength depends on many factors; probe labeling efficiency, the type of probe and the type of dye affect the fluorescent signal. Fluorescently-tagged antibodies or streptavidin are bound to the dye molecule. These secondary components are selected so that they have a strong signal.
Fibre FISH:
An alternative to inter-phase or metaphase preparations, fibre FISH, inter-phase chromosomes are attached to a slide in such a way that they are stretched out in a straight line, rather than being tightly coiled, as in conventional FISH, or adopting a random conformation, as in inter-phase FISH. This is accomplished by applying mechanical shear along the length of the slide; either to cells which have been fixed to the slide and then lysed, or to a solution of purified DNA.
A technique known as chromosome combing is increasingly used for this purpose. The extended conformation of the chromosomes allows dramatically higher resolution – even down to a few kilo-bases. The preparation of fibre FISH samples, although conceptually simple, is a rather skilled art, and only specialized laboratories use the technique routinely.
The incentive for multiple labeling of chromosomes and subsequent detection in multiple colors has given rise to Multi-color FISH (M-FISH), fuelled by the desire to maximize the efficiency of the FISH process when applied to situations such as the study of the complex translocations often found in cancer cytogenetics.
It is possible to detect multiple targets with fewer fluorophores (e.g., 3 fluors for 7 targets) based on labeling the probes combinatorially, with more than one fluor in various ratios. To simultaneously discriminate all 24 human chromosomes, 5 fluorochromes are required, each fluorescing in a distinct spectral region.
Complex “acquired” chromosome abnormalities are common in cancer cytogenetics, ranging from amplification of chromosome number to various defined translocations. One technique which is particularly suited to study of tumour DNA is Comparative Genomic Hybridisation (CGH), a development of chromosome painting, which detects a deviation of the ratio of two fluoro-chromes to indicate amplification or deletion along the chromosome strand. FISH can also be instrumental for gene mapping.
ADVERTISEMENTS:
Genes and DNA segments are continually being assigned to chromosome bands, but it is often necessary to determine the precise arrangement of each sequence relative to another at a more detailed level. Genomic in situ hybridization (GISH) is becoming an important technique to characterize parental genomes in wide hybrids and to detect introgressed segments and chromosome rearrangements. Total genomic DNA as a probe in situ hybridization has been used to detect parental chromosomes in wheat-rye hybrids and in discriminating closely related Triliceae species.
In situ hybridization using genomic probes has been demonstrated to be an effective technique for the detection of meiotic pairing between wheat-rye. However, GISH has not been used to characterize meiotic pairing in Oryza.
Technique # 2. Blotting Techniques:
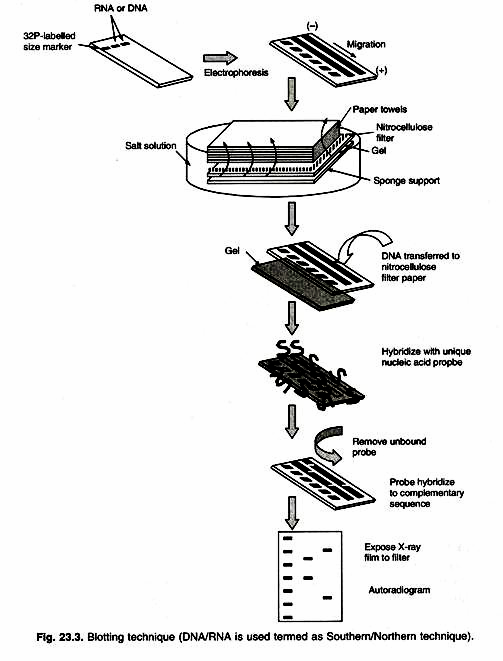
The term “blotting” refers to the transfer of biological samples from a gel to a membrane and their subsequent detection on the surface of the membrane.
Southern Blotting:
Southern blotting uses DNA molecule as marker (developed by E.M. Southern). Restriction enzyme digestion of genomic DNA results in so many fragments that a stained electrophoretic gel shows a smear of DNA. A probe can identify one fragment in this mixture, with the use of the technique called Southern Blotting.
ADVERTISEMENTS:
After DNA segments are fractionated on the gel, one absorbent membrane is laid on the gel and DNA bands are transferred (blotted) on to the membrane by capillary action. These DNA bands remain in the same relative position on the membrane.
The membrane is emerged in a labeled probe (a defined radio-labeled nucleic acid segment that is used to identify specific DNA molecules bearing the complimentary sequence, produced by PCR or traditional cloning method). An autoradiogram is used to demonstrate the presence of any bands on the gel that are homologous to the probe (Fig. 23.3). The cloned DNA finds widespread application as a probe for detecting either a specific clone or a specific DNA/RNA fragment. In all these cases the ability of nucleic acids to find and bind to complementary sequences in solution is being exploited.
Northern Blotting:
Northern blotting uses mRNA as marker. The southern blotting can be extended to detect a specific RNA molecule from a mixture of RNAs fractioned on a gel. This technique is called Northern Blotting to contrast it with the Southern’s technique for DNA analysis.
The fractioned RNA is blotted on to a membrane and probed in the same way as for Southern blotting. One application of Northern analysis is to determine whether specific gene is transcribed in certain tissue or under certain environmental conditions. RNAse is extracted from appropriate cell sample, then electro-phoresed, blotted and probed with the cloned gene in question. A positive signal shows presence of the transcript (Fig. 23.3).
Western Blotting:
Western blotting uses protein as marker compound. Western blotting (also called immune-blotting because an antibody is used to specifically detect its antigen) was introduced by Towbin et al. in 1979 and is now a routine technique for protein analysis. The specificity of the antibody-antigen interaction enables a single protein to be identified in the midst of a complex protein mixture. Western blotting is commonly used to positively identify a specific protein in a complex mixture and to obtain qualitative and semi-quantitative data about that protein.
The first step in a Western blotting procedure is to separate the macromolecules using gel electrophoresis. Following electrophoresis, the separated molecules are transferred or blotted onto a second matrix, generally a nitrocellulose or polyvinylidene fluoride (PVDF) membrane. Next, the membrane is blocked to prevent any nonspecific binding of antibodies to the surface of the membrane. The transferred protein is complexes with an enzyme-labelled antibody as a probe.
An appropriate substrate is then added to the enzyme and together they produce a detectable product such as a chromogenic or fluorogenic precipitate on the membrane for colorimetric or fluorimetric detection, respectively. The most sensitive detection methods use a chemiluminescent substrate that, when combined with the enzyme, produces light as a byproduct. The light output can be captured using film, a CCD camera or a phosphor imager that is designed for chemiluminescent detection.
Whatever substrate is used, the intensity of the signal should correlate with the abundance of the antigen on the blotting membrane.
ADVERTISEMENTS:
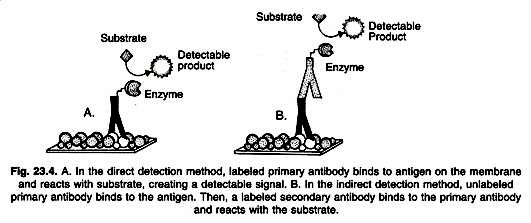
Detailed procedures for detection of a Western blot vary widely. One common variation involves direct vs. indirect detection as shown in Figure 23.4. With the direct detection method, the primary antibody that is used to detect an antigen on the blot is also labeled with an enzyme or fluorescent dye.
In the indirect detection method, a primary antibody is added first to bind to the antigen. This is followed by a labeled secondary antibody that is directed against the primary antibody. Labels include biotin, fluorescent probes such as fluorescein or rhodamine, and enzyme conjugates such as horseradish per-oxidase or alkaline phosphatase. The indirect method offers many advantages over the direct method.
Protein Transfer to a Membrane:
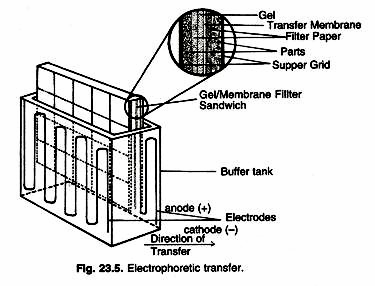
Following electrophoresis, the protein must be transferred from the electrophoresis gel to a membrane. There are a variety of methods that have been used for this process, including diffusion transfer, capillary transfer, heat-accelerated convectional transfer, vacuum blotting transfer and electro elution. The transfer method that is used most commonly used for proteins is electro elution or electrophoretic transfer, because of its speed and transfer efficiency. This method uses the electrophoretic mobility of proteins to transfer them from the gel to the matrix.
Electrophoretic transfer of proteins involves placing a protein-containing polyacrylamide gel in direct contact with a piece of nitrocellulose or other suitable, protein-binding support and “sandwiching” this between two electrodes submerged in a conducting solution (Fig. 23.5). When an electric field is applied, the proteins move out of the polyacrylamide gel and onto the surface of the membrane, where the proteins become tightly attached. The resulting membrane is a copy of the protein pattern that was found in the polyacrylamide gel itself.
Transfer efficiency can vary dramatically among proteins, based upon the ability of a protein to migrate out of the gel and its propensity to bind to the membrane under a particular set of conditions. The efficiency of transfer depends on factors such as the composition of the gel, complete contact of the gel with the membrane, the position of the electrodes, the transfer time, size and composition of proteins, field strength and the presence of detergents. Optimal transfer of proteins is generally obtained in low ionic strength buffers and with low electrical current.
ADVERTISEMENTS:
The most commonly used membranes for Western blotting include nitrocellulose, polyvinylidene difluoride (PVDF) and nylon membranes. After transfer and before proceeding with the Western blot, it is often desirable to stain all proteins on the membrane with a reversible stain to check the transfer efficiency. Although the gel may be stained to determine that protein left the gel, this does not ensure efficient binding of protein on the membrane. Ponceau S stain is the most widely used reagent for staining proteins on a membrane. However, it has limited sensitivity, does not photograph well, and fades with time.
MemCode Reversible Stain is a superior alternative for staining protein on nitrocellulose or PVDF membranes. MemCode Stain detects low nanogram levels of protein, is easily photographed, does not fade with time and takes less than 30 minutes to stain, photograph and erase.
Blocking Nonspecific Binding Sites:
In a Western blot, it is important to block the un-reacted sites on the membrane to reduce the amount of nonspecific binding of proteins during subsequent steps in the assay. A variety of blocking buffers ranging from milk or normal serum to highly purified proteins have been used to block un-reacted sites on a membrane. The blocking buffer should improve the sensitivity of the assay by reducing background interference.
Individual blocking buffers are not compatible with every system. For this reason, a variety of blockers in both Tris buffered saline (TBS) and phosphate buffered saline (PBS) are available. The proper choice of blocker for a given blot depends on the antigen itself and on the type of enzyme conjugate to be used. For example, with applications using an alkaline phosphates conjugate, a blocking buffer in TBS should be selected, because PBS interferes with alkaline phosphates.
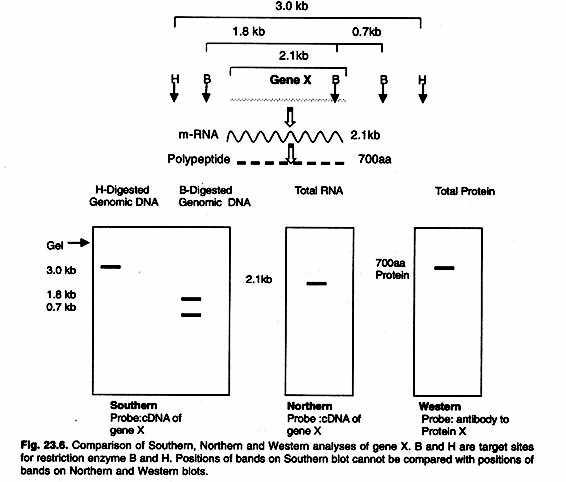
The ideal blocking buffer will bind to all potential sites of nonspecific interaction, eliminating background altogether without altering or obscuring the epitope for antibody binding. A complete line of blocking buffers for Western blotting includes BLOTTO, Casein, BSA, SEA BLOCK and the exclusive Super-Block Blocking Buffers. A comparison of three blotting techniques is given in the Figure 23.6.
Washing:
Like other immunoassay procedures, Western blotting consists of a series of incubations with different immunochemical reagents separated by wash steps. Washing steps are necessary to remove unbound reagents and reduce background, thereby increasing the signal: noise ratio.
Insufficient washing will allow high background, while excessive washing may result in decreased sensitivity caused by elution of the antibody and/or antigen from the blot. As with other steps in performing a Western blot, a variety of buffers may be used.
Occasionally, washing is performed in a physiological buffer such as Tris buffered saline (TBS) or phosphate buffered saline (PBS) without any additives. More commonly, a detergent such as 0.05% Tween 20 is added to the buffer to help remove nonspecifically bound material.
Primary and Secondary Antibodies:
The choice of a primary antibody for a Western blot will depend on the antigen to be detected and what antibodies are available to that antigen. A huge number of primary antibodies are available commercially and can be identified quickly by searching sites. Both polyclonal and monoclonal antibodies work well for Western blotting.
Polyclonal antibodies are less expensive and less time-consuming to produce and they often have a high affinity for the antigen. Monoclonal antibodies are valued for their specificity, purity and consistency that result in lower background. A wide variety of labeled, secondary antibodies can be used for Western blot detection.
The choice of secondary antibody depends upon the species of animal in which the primary antibody was raised (the host species). A wide variety of secondary antibodies used in Western blotting are biotin, fluorescein, rhodamine, horseradish peroxidase and alkaline phosphatase.
Technique # 3. Polymerase Chain Reaction:
A powerful technique for amplifying DNA was discovered by Kary Mullis of Cetus Corporation, USA in 1985. The prestigious journal ‘Science’ published Polymerase Chain Reaction (PCR) as major scientific development and Taq DNA polymerase enzyme used in the technique was described as ‘molecule of the year’ in 1989.
PCR is so powerful that from single copy of DNA molecule, millions of copies can be obtained with high accuracy, specificity and in very short time. So in few years since its invention, the polymerase chain reaction (PCR) has become a convenient and trustworthy alternative to gene cloning using vectors.
Mechanism of Basic PCR:
PCR can be defined as recurring amplification of a particular region of the genome preselected with the help of primers flanking the region of the genome to be amplified. The DNA replication involves polymerization of nucleotides, using template DNA strand with the help of enzyme DNA polymerase and suitable primers.
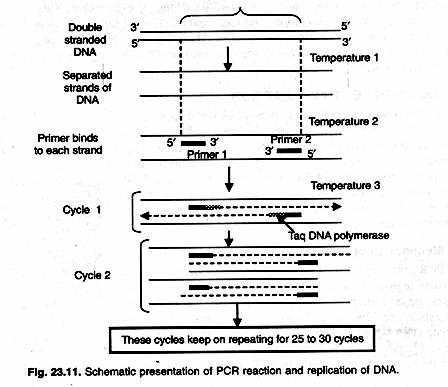
In living cells small single stranded RNA molecule acts as primer, while in PCR short DNA molecules (which are complementary to two sites- either side of the piece of DNA to be amplified) are added as primers. Primers added are usually synthesized oligo-nucleotides. In PCR this reaction takes place in an ‘eppendorf tube’ instead of living cells (Fig. 23.10).
The purpose of a PCR (Polymerase Chain Reaction) is to make a huge number of copies of a gene. This is necessary to have enough starting template for sequencing.
The Cycling Reactions:
There are three major steps in a PCR, which are repeated for 30 or 40 cycles in order to obtain large number of copies of desired DNA template (Fig. 23.11). This is done on an automated cycler, which can heat and cool the eppendorf tubes with the reaction mixture in a very short time.
PCR or Thermal Cycler Machine:
1. Denaturation at 90-98 °C:
During the denaturation, the double strands of DNA separate. All enzymatic reactions stop (for example: the extension from a previous cycle). Higher temperature may be required to denature template and/or target DNA that are rich in G+C (>55%). For routine amplification of linear DNA template whose G+C is 55% or less denaturation is done at 94 °C for 45 seconds.
2. Annealing at 40-60 °C:
Temperature 54°C anneals two complementary primers to ends of separated single-strands of target DNA. The primers are jiggling around, caused by the Brownian motion. Ionic bonds are constantly formed and broken between the single stranded primer and the single stranded template.
The more stable bonds last a little bit longer (primers that fit exactly) and on that little piece of double stranded DNA (template and primer); the polymerase can attach and starts copying the template. Once there are a few bases built in, the ionic bond is so strong between the template and the primer, that it does not break anymore.
3. Extension at 72 °C:
This is the ideal working temperature for the polymerase. At this temperature thermostable Taq DNA polymerase use single stranded DNA strands of target and primers to synthesize new strands. The primers have a strong ionic attraction to the template than the forces breaking these attractions.
Primers that are on positions with no exact match get loose again (because of the higher temperature) and don’t give an extension of the fragment. The bases (complementary to the template) are coupled to the primer on the 3′ side (the polymerase adds dNTP’s from 5′ to 3′, reading the template from 3′ to 5′ side; bases are added complementary to the template).
This three temperature cycle is repeated 25-30 times and the result is million -fold increase of the copies of target DNA. Reaction is done in DNA thermal cycler using step cycle program set to denature at 94 °C for 45 sec, anneal at 54 °C for 20 sec and extend at 72 °C for 30 sec.
Earlier DNA polymerase enzyme from E.coli was used. But as it is sensitive to heat as heat destroyed the enzyme new enzyme had to be added. Use of thermostable DNA polymerase enzyme of Thermophilus aquaticus (from hot springs) was reported by Saiki and co-workers in 1988 and that was the major breakthrough in PCR development.
It could give more yields, could avoid repeated addition of enzyme, was highly specific and longer fragments up to 10 kb can be amplified instead of less than 400bp earlier. This enzyme act best at 72 °C and the denaturation of 90 °C does not destroy its enzymatic activity. Other thermostable DNA polymerase enzymes used today in PCR are Pfu DNA polymerase from Pyrococcus furiosus, Vent polymerase from Thermococcus litoralis and Tbr polymerase from T. brockianus etc and they are found to work more efficiently.
Comparison of PCR with Gene Cloning:
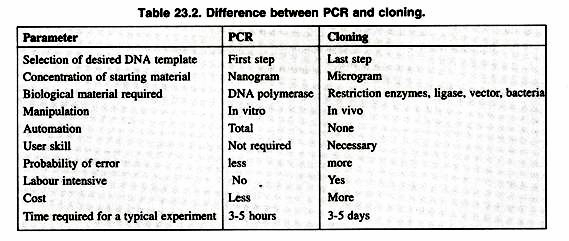
PCR will ultimately replace gene cloning to a large extent except some applications of gene cloning like production of a protein in large quantities for human use. The final result of both gene cloning and PCR can be said to be a selective amplification of specific sequences. Some major points of comparison between the both are as shown in Table 23.2.
Applications:
1. PCR for screening of uncharacterized mutations:
Small insertions or deletions can be simply detected by designing primers from regions closely flanking the mutation site and distinguishing the normal and mutant alleles by size on polyacrylamide or agarose gels.
If the mutation changes a restriction site, mutant and normal alleles can be distinguished by amplifying across the mutant site and digesting the PCR product with relevant restriction endo-nuclease.
2. Isolation of genomic DNA:
PCR allows isolation of DNA fragments from genomic DNA by selective amplification of a specific region of DNA. This use of PCR augments many methods, such as Southern and Northern blotting and DNA cloning that require large amounts of DNA, representing a specific DNA region. PCR supplies these techniques with high amounts of pure DNA, enabling analysis of DNA samples even from very small amounts of starting material.
3. Amplification and quantification of DNA:
Because PCR amplifies the regions of DNA that it targets, PCR can be used to analyze extremely small amounts of sample. This is often critical for forensic analysis, when only a trace amount of DNA is available as evidence. PCR may also be used in the analysis of ancient DNA that is thousands of years old. These PCR-based techniques have been successfully used on animals, such as a 40,000 years old mammoth, and also on human DNA. Viral DNA can likewise be detected by PCR.
4. Human Health and the Human Genome:
PCR has very quickly become an essential tool for improving human health and human life. Medical research and clinical medicine are profiting from PCR mainly in two areas: detection of infectious disease organisms, and detection of variations and mutations in genes, especially human genes. The primers used need to be specific to the targeted sequences in the DNA of a virus, and the PCR can be used for diagnostic analyses or DNA sequencing of the viral genome.
The method is especially useful for searching out disease organisms that are difficult or impossible to culture, such as many kinds of bacteria, fungi, and viruses, because it can generate analyzable quantities of the organism’s genetic material for identification. In short, if a disorder is caused by an infectious agent, PCR can, in principle, search out the culprit. More than 60 PCR protocols for identifying pathogens have been described to date.