ADVERTISEMENTS:
In this article we will discuss about:- 1. Structure of Streptomycin 2. Antibiotic Spectrum of Streptomycin 3. Mechanism of Action 4. Dependence.
Structure of Streptomycin:
Streptomycin is characterised chemically as an aminoglycoside antibiotic. It consists of three components linked glycosidically (by ether bonds):
(i) Streptidine (inositol with two guanido groups),
ADVERTISEMENTS:
(ii) Streptose (methyl pentose), and
(iii) Streptoscamine (N-methyl-L-glycosamine) as shown in Fig. 45.9.
Both guanido groups of streptidine are essential for the antibiotic activity and removal of one group reduces antibiotic activity upto 90%.
Antibiotic Spectrum of Streptomycin:
Streptomycin is bactericidal and broad-spectrum antibiotic. It is active against both gram-positive and gram-negative bacteria. Streptomycin is inhibitory for several species of Mycobacterium and is an effective antibiotic for treatment of tuberculosis caused by M. tuberculosis.
ADVERTISEMENTS:
Streptomycin is toxic to humans and other animals and causes side effects such as allergic responses, loss of hearing, nausea, and kidney damage. Highly purified streptomycin is nontoxic when given in small doses, but it appears to have a cumulative detrimental effect on nervous system when, given as a medication over long periods of time.
Mechanism of Action of Streptomycin:
Streptomycin, like other aminoglycosidic antibiotics (e.g., gentamycin, neomycin, kanamycin, tobramycin), inhibits protein synthesis in bacterial cells by binding to the 30S subunit of ribosomes.
By doing so, the streptomycin causes a structural change which interferes with the recognition site of codon-anticodon interaction resulting in misreading of the genetic message carried by messenger RNA (mRNA). The mechanism of inhibition of protein synthesis by streptomycin is schematically shown in Fig. 45.10.
Streptomycin-Dependence:
It has been found that streptomycin-susceptible bacteria may become resistant to the antibiotic by a single step mutation. Such mutation results in a structural alteration in the 30S subunit leading to a loss of affinity for streptomycin, so that the antibiotic can no longer bind its recognition site and inhibit protein synthesis.
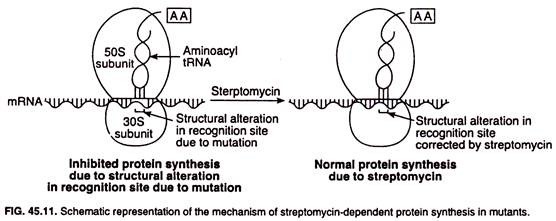
A very interesting observation has been made in some streptomycin-resistant mutants and is called streptomycin- dependence. Streptomycin-dependence is thought to develop as a result of another mutation which causes an opposite distortion of the recognition site of the 305 subunit of ribosome.
In streptomycin-dependent mutants of bacterial pathogen, protein synthesis fails to take place in absence of streptomycin. The schematic representation of the mechanism of streptomycin dependence is shown in Fig. 45.11.