ADVERTISEMENTS:
The below mentioned article provides notes on conjugation.
Note # 1. Role of Surface Protein in Conjugation:
Conjugation differs from transformation in the fact that in the former physical contact is established between two different strains through a conjugation tube. The genetic material from the donor cell (male) is transferred to the recipient (female) cell. There are special appendages present on bacterial cell surface which are called sex pilus or F pilus which forms the conjugation tube.
The fertility (F) factor enables the cell to act as donor. The F factor of donor cell includes the information’s of sex pili the number of which varies from 1 to 3. The cells containing an autonomous F are referred to as F+ cells. It replicates independently. There are only 1-3 copies of F factor per cell.
ADVERTISEMENTS:
Under certain specific conditions the number of pili per cell goes to five. The number of pili corresponds to the number of copies of F factor. This explains that each F factor synthesizes a single pilus whether it is autonomous replicating conditions (as plasmid) or in integrated conditions (as episome).
Moreover, the recipient cells possess receptor sites on cell surfaces which are required for conjugation. Certain bacteriophages e.g. f2, MS2, and Qβ act as donor. The donor E. coli cells possess sex pili as well as type I pilus on their cell surfaces. For example phage M12 is adsorbed randomly only on sex pili but not on cell surfaces of recipient bacterial cell.
Note # 2. The F Factor:
The presence of F factor in a bacterial cell determines its autonomous replication, sex pili formation and conjugal transfer function. Thus, it governs the sexuality and conjugation.
Two mating types in E. coli K12 have been found to depend on presence and absence of the F factor. The F factor remains in two stages as plasmid and as episome. The F plasmid replicates independently. However, sometimes it is integrated with the normal chromosome of the bacterium. Therefore, it is referred to as episome.
ADVERTISEMENTS:
The F factors have shown the following significant features:
(a) When F+ strain of a bacterium is incubated on a nutrient medium mixed with acridine orange, it is converted into F– strain. Acridine orange is effective only with the growing bacteria as it inhibits the autonomously replicating F factor.
(b) A cross between two F– strains does not yield recombinants. It is always sterile as the F– strain cannot undergo conjugation with the other F– strain.
(c) The crosses between F+ and F+ strain yield F+ cells but a very low level. Some of F+ cells are converted into F– genotype.
(d) Transfer of F factor from F+ to F– in F+ x F– crosses occurs at a frequency of about 100% but the production of recombinants occurs at the rate of one per 104 to 105 cells.
(e) Among F+ strains there are certain F+ sub-strains that show about 1000 time more rate of recombination with F– strains. The sub-strains are called high frequency recombination (Hfr) strains. The Hfr strains are produced when F factor integrates with the bacterial chromosomes.
Note # 3. Genetic Map of F Plasmid:
A genetic map of F plasmid is shown in Fig. 8.9. The F plasmid of E. coli is about 100 kb with genes coding for autonomous replication, sex pili formation and conjugal transfer function. The F plasmid contains the transfer (tra) region and non-transfer related markers.
ADVERTISEMENTS:
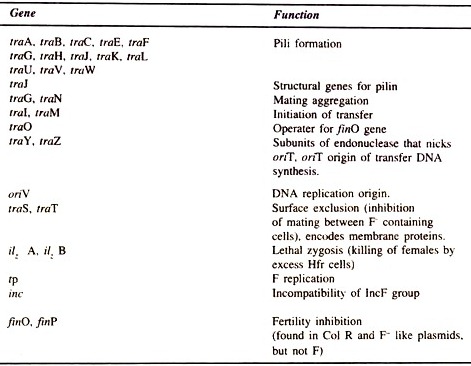
The non-transfer related markers are the insertion sequences (IS3, δ, ү and IS2), stable DNA degradation (srn B), inhibition of replication by T7, and II phages (pif), and a region for replication (rep), incompatibility (inc) and origin of vegetative replication (oriV). Some genes of R plasmid with their functions are given in Table 8.1.
Willetts and Wilkins (1984) have given the physical and genetic map of transfer region of F plasmid (Fig. 8.10) which is about 32 kb long consisting of about 25 known transfer genes.
Twelve genes are involved in F pilus formation (e.g. tra A,-L,-E,-K,-B,-V,-W/C,-U,-F,-H,-G). Genes involved in regulation are finP and traJ. Stabilization of mating pairs is done by genes traN and traG, conjugative DNA metabolism by traM, traY, traD, tral and traZ and surface exclusion by traS and traT.
Fig. 8.10 : Physical and genetic map of the transfer (tra) region of F-plasmid.
ADVERTISEMENTS:
The traM and traJ promoter regions have been sequenced and traY-Z operon possesses its own promoter. Transcription from the promoters for traM and traY-Z operon is dependent on the product or traJ which in turn is negatively regulated by the FinOP repressor. The tral and traZ genes are transcribed continuously from a second promoter at about 18% or the level from the tral induced traY-Z operon promoter.
Note # 4. The Conjugal Transfer Process:
In the Enterobacteriaceae specific structural appendages i.e. sex pili are found on cell surfaces of donor cell. The donor cells contain F factor. Cell-to-cell contact between F+ and F– is established. The steps of F plasmid transfer from F+ to F– cell are shown in Fig. 8.11 A-E.
A pool of preformed subunits is incorporated into mature sex pili. The number of sex pili vary from 1 to 3 per cell. The tip of pilius is involved in the stable mating pair formation (governed by traN and troF genes) when interacts with the ompA gene product on the outer surface of the recipient cell.
ADVERTISEMENTS:
After the initial contact between the tip of pilus and recipient cell (A) the pilus contracts and brings the F+ and F– cells into the close proximity (B). This wall to wall contact forms a conjugation bridge involving the fusion of the cell envelopes (Figs. 8.11B and 8.12). At oriT site of plasmid a nick is made by traYZ endonuclease yielding in 5′-terminus single strand that invades the recipient cells.
The 5′-terminus of DNA binds with a pilot protein and travels gradually through this membrane bridge (probably a pore involving the traD DNA gene product (Fig. 8.12) but not through the pilus itself as it was originally believed. It has been found that the mating mixture of E. coli form mating aggregates of 2- 20 cells each rather than only mating pairs.
After the formation of mating aggregates transfer of F+ DNA starts from oriF region as opposed to oriV as a plasmid enclosed endonuclease (tral gene product) nicks the F plasmid at oriT. The intact strand acts as template and the 5′ end strand is transferred to the recipient cell through a rolling circle mechanism of replication (C-D).
Fig. 8.12 : A model for conjugative transfer at F plasmid.
At the nicked oriT site, the traM triggers conjugal DNA synthesis by exposing sufficient ssDNA to facilitate the binding of helicase (a tral gene product) or DNA helicase I (Fig.8.12). The helicase I move on the other strand which is under going transfer for unwinding the plasmid duplex. Helicase I migrates with DNA polymerase III synthesizing the replacement strand of the donor DNA.
If the helicase I binds to the membrane complex during conjugation, the concomitant ATP synthesis might provide the motive force to displace the transferred strand into the recipient cell. After entering into the recipient cell, the 5′ end strand is attached to the membrane and undergoes replication.
Transfer of DNA is associated with synthesis of a replacement strand in the donor cell and of a complementary strand in the recipient cell. Both the processes require de novo primer synthesis and the activity of DNA polymerase III holoenzyme.
It is assumed that a single strand binding protein coats the DNA and helps the conjugal DNA synthesis. This depends upon the nature of the pore. This protein is also transferred from the donor to the recipient cells. After the synthesis of complementary strand the F plasmid is circularized (Fig. 8.11 E).
Note # 5. Barriers to Conjugation:
It has been found that sometimes the cells containing F factor are poor recipient, when conjugative crosses occur. This is due to the presence of surface exclusion. A similar phenomenon (incompatibility) occurs when a F’ element is transferred into a recipient cell that already contained F plasmid.
ADVERTISEMENTS:
The F’ element renders F plasmid to become unable for fertility. It has been found out that for surface exclusion two genes (traS and traT) are required with traT protein which is an outer membrane-protein. It is hoped that tral protein may block the stabilization sites of mating pair or inhibits the structural proteins required for stabilization of mating pair.
Table 8.1 : Some genes and sites of F plasmids and their function.
In addition, in most of the conjugative plasmids e.g. R 100 transfer of DNA is markedly reduced as compared with F. This is because the fertility inhibition system (FinOP) controls the regulatory system of tra genes. The finO and finP gene products interact and form a FinOP inhibitor of tra gene expression.
Note # 6. The High Frequency Recombination (Hfr) Strains:
When the chromosome of F+ cell integrates with F plasmid, it is called high frequency recombination (Hfr) cell. The Hfr cells arise from F+ cultures (Fig.8.13A). Even after integration of F into chromosome, the chromosome retains a single, circular DNA molecule. The F acts as it was a part of the chromosome.
The F increases the size of chromosome. However F is capable of transferring the whole chromosome from Hfr cells to the F– culture. The frequency of insertion occurs at about 105 – 107 per generation i.e. in a bacterial population of 107 F+ cells; there is possibility of 1-100 cells in having an integrated F plasmid with chromosome.
ADVERTISEMENTS:
Following are some of the differences between F+ cells and Hfr cells:
(a) The F factor of Hfr cells is rarely transferred during recombination. In an Hfr x F, the frequency of recombination is high and that of transfer of F factor in low. In contrast in F+ X F– cross, the frequency of recombination is very low and that of transfer of F factor is high.
(b) It takes about 2 minutes for transfer of F, whereas 100 minutes for entire bacterial chromosome transfer. This difference is mainly due to the relative size of F and the integrated chromosome.
(c) In a cross between F– and Hfr cells, F– cells always remain F– because of separation of cells before final transfer of ultimate F segment.
(d) In a mating between an Hfr leu+ culture and an F– leu– culture, F– leu+ cells arise. The genotype of the donor is not changed because the concurrent replication in the donor replaces the transferred DNA strand.
(a) Mechanism of integration:
The mechanism of integration is shown in Fig. 8.13B. Integration is a reciprocal DNA exchange. From base sequencing study it is clear that an integrated F sequence is flanked by two copies of one of the insertion sequences (IS elements) present in F plasmid. Thus, integration involves homologous recombination between two circular DNA molecules resulting in one circular molecule that contains both the DNAs.
Both the IS elements (IS2 and IS3) present on F plasmid and IS element on bacterial chromosome (E. coli also contain 5 each of IS2 and IS3 sequences) set as the homologous regions for insertion. The integration of F plasmid depends on recA but rarely independent on recA. The F integration also takes place depending on transposition of IS elements.
Thus Hfr cells arise due to homologous recombination between the two identical IS elements; one is present in chromosome and the other in F plasmid. Secondly, the Hfr cells also arise by forming a co-integrate mediated by an IS element in F plasmid, and by duplicating a target sequence in the chromosome.
There is directionality to conjugal DNA transfer. The direction of transfer from oriT is such that the tra genes are transferred in the last. The orientation of chromosomal IS element is such that the host gene B is as a proximal marker, whereas gene A is transferred in the last.
After integration, the F DNA replicates along with the host chromosome. When the host dnaA gene is non-functional, replication of whole chromosome can begin from an integrated F DNA.
(b) The order of chromosome transfer and conjugation mapping:
Wollman (1956) determined, by interrupted mating experiments, the order of chromosome transfer from an Hfr donor to an F– recipient cell. The time at which a particular gene enters a recipient is related to the portion of the genes on the chromosome. A map can be obtained from the time of entry of each gene.
The interrupted mating experiment involves:
(i) Mixing of an Hfr strain with F– strain,
(ii) Interrupting the conjugation at certain intervals by breaking the cells apart in a high speed blender,
(iii) Plating the cells on various types of selective media to select the recombinant cells that had received the genes from Hfr strain before interruption of mating.
A simplified linkage map of circular E. coli chromosome constructed from interrupted mating experiment is shown in Fig. 8.14.
It takes about 100 minutes to transfer the entire E. coli chromosome. Therefore, the genes are mapped relative to the position of the integrated F plasmid by determining the time taken by the gene to be transferred to the recipient cell.
The interrupted mating experiment also reveals the frequency of recombination of each marker identified by detectable mutation at a particular locus (Fig. 8.14). The genetic markers are leu, lac and gal. The donor Hfr cell is wild type, whereas the recipient is leu, lac–, gal– i.e. the recipient is mutant in lac and gal but wild type in leu.
Therefore, the mutant requires lactose or galactose as carbon source. The Hfr is streptomycin- sensitive (Strs) and the recipient is streptomycin- resistant (Strr). After mixing the donor and recipient cells at zero time, the aliquots of mixture are removed at different intervals and mating pairs disrupted by blending.
The mixture is plated on minimal media containing:
(i) Glucose to select for Leu+ recombinant,
(ii) Lactose plus leucine to select for Lac+ recombinants, and
(iii) Galactose + leucine to select for Gal+ recombinants.
Colonies growing on these media are the recombinants i.e. the recipient into which the wild type donor gene was transferred and replaced the mutant gene.
Thus, the conjugate transfer of Hfr chromosome is time dependent. Each gene enters the F– cell at a particular time. By measuring different time intervals a graph can be plotted (Fig. 8.14) and linkage map can be constructed (Fig. 8.15).
(c) High Resolution Mapping:
One gets low resolution mapping by interrupted mating experiment. Moreover, this method is not useful for high resolution mapping within a distance of 2 minutes. Hence, study of stable recombinants rather than the gene transfer is required.
For example, if abc+ and thr+ are the two genes transferred, the frequency of colony can be calculated with thr+ and thr– among those with abc+ genes if abc+ is more frequent than thr+.
The recombinational distance between abc+ and thr+ can be obtained from the proportion of thr– (abc+ thr–) among total abc+ i.e. [(abc+ thr+) + (abc+ thr–)] as below:
Recombination distance = (abc+ thr–)/(abc+ thr+) + (abc+ thr–)
Note # 7. Formation of F–Prime (F’):
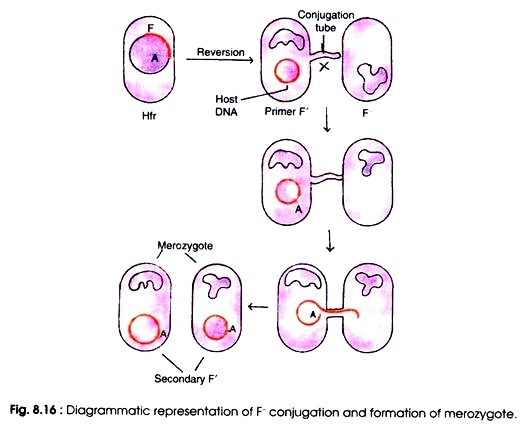
Integration of F factor is a reversible process. The Hfr cell can revert to the F+ state again. When the reversible process occurs the F factor is set free from the chromosome and resumes its autonomously replicating capability. Separation of F factor from the integrated chromosome occurs aberrantly at a low frequency and yields plasmid containing F factor and a small segment of chromosome is called F’ cells.
The F’ is of two types. Type I F’ has lost some sequence but carries some host DNA located at one or the other side of the integrated F. Type II F’ contains all of F’ plus some host DNA from both sides of the point where F was integrated. In both the condition F contains a small segment of chromosome.
When such primary F’ cells are crossed with F– recipients, the F factor is transferred efficiently together in F– converting them into the secondary F’ cells.
The secondary F cells are partially diploid hence called as merodiploid or merozygote because the recipient cells, in addition to its own chromosome, contained a segment of DNA from the donor cell i.e. F cell (Fig. 8.16). This process of transfer of bacterial DNA from donor cell to recipient cell as a part of sex factor has been called sexduction by Jacob and Wollman (1961).
The F’ conjugation is very important in the study of microbial genetics to find out whether the allele carried by an F’ plasmid in merozygote is dominant or recessive to the chromosomal gene. Study of F’ plasmid is also useful in mapping the chromosome since the two neighbour genes are picked up by an F factor.