ADVERTISEMENTS:
In this article we will discuss about the meaning of tRNA.
The genetic message contained in the nucleotide sequence of transcribed RNA cannot be read off directly by the amino acids in the polypeptide chain. Crick and Hoagland were first to postulate that an intermediate RNA molecule functions as an “adaptor” into which the amino acid is plugged so that it can be adapted to the nucleotide triplet language of the genetic code.
When first discovered it was given the name soluble RNA (sRNA), but later it was called transfer RNA (tRNA) which described its function more accurately.
ADVERTISEMENTS:
A particular tRNA first attaches to its specific amino acid and transports it to the ribosome against a specific nucleotide sequence on the mRNA. The tRNAs for 20 different amino acids all have different structures, and each is specifically adapted for H-bonding with the nucleotide sequences in mRNA.
Each amino acid has at least one corresponding tRNA and some have multiple tRNAs. For example there are 5 distinctly different tRNAs specific for leucine in E. coli cells. Moreover there are different types of tRNAs for a given amino acid present in mitochondria and in the cytoplasm of eukaryotic cells.
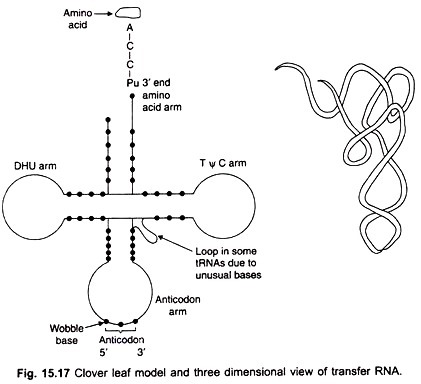
The tRNA molecules are single-stranded, have about 73 to 93 nucleotides, a molecular weight of about 25,000 and sedimentation coefficient of about 4S. The single-stranded molecules can fold on themselves forming hairpins and many of the bases become linked by H-bonds.
The adenine residues pair with uracils and guanines with cytosines. Due to this intrachain base pairing about 60-70 per cent of the tRNA structure exists in double stranded form. Although tRNAs for different amino acids have quite different base compositions, yet due to folding and intrachain base pairing, all of them basically form a four-leaved clover-like structure (rarely three-leaved as in alanyl-tRNA). The single-stranded regions in tRNA are represented by loops (Fig 15.17).
In higher organisms the genes for tRNA are reiterated (repeated). Thus bacteria have only 1 or 2 copies of each tRNA gene, yeasts have 5-7 copies, Drosophila has 13 copies, and Xenopus and mammalian cells about 200. In eukaryotes, precursors to tRNAs reach the cytoplasm; they are longer than tRNA by about 30 nucleotides.
Within an hour after reaching the cytoplasm they are cleaved to the dimensions of tRNA and some nucleotides become modified by methylation. Some of the uridine residues change into pseudouridine and dihydrouridine residues. Most tRNAs have many unusual bases such as inosine (I), pseudo-uracil (ψ) and some methylated forms of normal bases.
The first complete nucleotide sequence of a tRNA was established by Holley and his colleagues in 1965 for yeast alanine tRNA. This was rewarded by a Nobel Prize 3 years later. All the tRNAs have the same terminal sequence CCA at the 3′ end of the amino acid arm.
The last residue adenylic acid is the site to which the aminoacyl group is esterified by the enzyme aminoacyl synthetase. Besides the CCA terminus, tRNAs have three loops.
The first TC loop of all tRNAs contains the pseudouridine (ψ) residue; in the second or dihydrouridine (DHU) loop, a dihydrouridine is present. The third is the anticodon loop containing a specific triplet of bases which is different in tRNAs for different amino acids. This triplet is also complementary to the corresponding codon triplets in mRNA with which it can form H-bonds.
The anticodon arm ensures that the correct amino acid is selected and placed on the ribosome for addition to a growing polypeptide chain. The third base of the anticodon occupying the 5′ position is the wobble base which is not so specific as the other two bases in its interaction with the corresponding base in the tRNA codon.
A tRNA contains at least two other specific sites, the enzyme recognition site, at which the tRNA is bound to the corresponding activating enzyme, and a second ribosome attachment site. The presence of unusual nucleotides is a striking feature of most tRNAs. They probably arise by enzymic modifications of the already existing bases. Since unusual bases cannot form base pairs, they lie exposed in the single stranded loops of the tRNA molecule.