ADVERTISEMENTS:
Read this article to learn about the concept and structure of operon mode in gene regulation.
Jacob and Monod (1961) gave the concept of Operon model to explain the gene regulation in prokaryotes using simple experiments to grow E. coli in petriplates containing histidine, lactose or lactose and glucose in the medium (Fig. 9.2).
When histidine is present in the medium, activity of histidine synthesizing enzymes decreases rapidly showing switching off the genes. Similarly, when concentration of lactose increases in the medium, activity of galactosidase increases.
ADVERTISEMENTS:
In a medium if both glucose and lactose are present, bacteria will utilize glucose first and there will be no β-galactosidase activity. Activity of β-galactosidase increases with the depletion of glucose and availability of lactose. Activity of P-galactosidase shut off if glucose is added in the medium. A molecule of lactose forms one molecule of galactose and one molecule of glucose.
These experiments clearly demonstrate substrate induced gene expression or inhibition. Therefore, a precise control mechanism must exist to switch on and switch off the genes. During 60s there was no clear vision about physical nature of gene even then they proposed gene regulation which holds true till today. Jacob and Monod were awarded Nobel Prize in 1965.
Structure of Operon:
ADVERTISEMENTS:
An Operon consists of a group of coordinately controlled genes that are all related to a particular cellular function. Operons occur primarily in prokaryotic organisms. Typically, the genes are physically adjacent to one another and all controlled from a single upstream promoter. An operator locus immediately adjacent to the promoter determines when the Operon is actually actively transcribed. In bacteria, genes are clustered into Operons: gene clusters that encode the proteins necessary to perform coordinated function, such as biosynthesis of a given amino acid.
RNA that is transcribed from prokaryotic Operons is polycistronic a term meaning that multiple proteins are encoded in a single transcript. In bacteria, control of the rate of transcriptional initiation is the predominant site for control of gene expression.
As with the majority of prokaryotic genes, initiation is controlled by two DNA sequence elements that are approximately 35 bases and 10 bases, respectively, upstream of the site of transcriptional initiation and as such are identified as the 35 and 10 positions. These 2 sequence elements are termed promoter sequences, because they promote recognition of transcriptional start sites by RNA polymerase.
The consensus sequence for the -35 position is TTGACA, and for the -10 position, TATAAT. (The -10 position is also known as the Pribnow- box). These promoter sequences are recognized and contacted by RNA polymerase (Fig. 9.3).
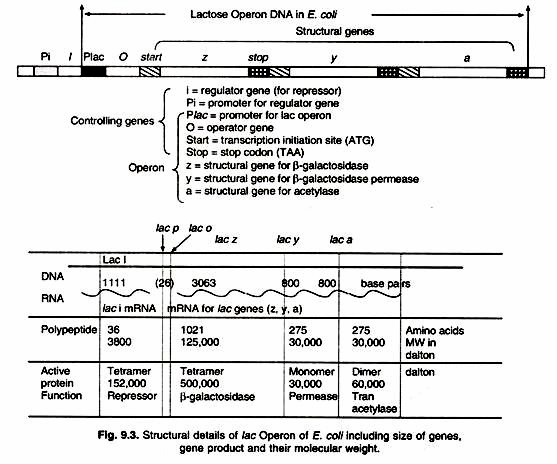 The lac Operon consists of one regulatory gene (the i gene) and three structural genes (z, y, and a). The i gene codes for the repressor of the lac operon. The z gene codes for β-galactosidase (3-gal), which is primarily responsible for the hydrolysis of the disaccharide, lactose into its monomeric units, galactose and glucose.The y gene codes for permease, which increases permeability of the cell to β-galactosides. The α gene encodes a transacetylase.
The lac Operon consists of one regulatory gene (the i gene) and three structural genes (z, y, and a). The i gene codes for the repressor of the lac operon. The z gene codes for β-galactosidase (3-gal), which is primarily responsible for the hydrolysis of the disaccharide, lactose into its monomeric units, galactose and glucose.The y gene codes for permease, which increases permeability of the cell to β-galactosides. The α gene encodes a transacetylase.
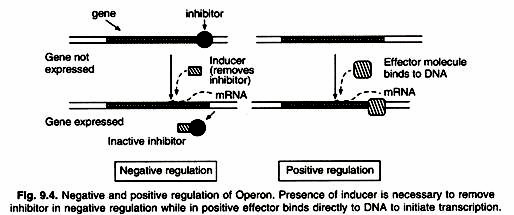
The activity of RNA polymerase at a given promoter is in turn regulated by interaction with accessory proteins, which affect its ability to recognize start sites. These regulatory proteins can act both positively (activators) and negatively (repressors) (Fig. 9.4). The accessibility of promoter regions of prokaryotic DNA is in many cases regulated by the interaction of proteins with sequences termed operators. The operator region is adjacent to the promoter elements in most Operons and in most cases the sequences of the operator bind a repressor protein.