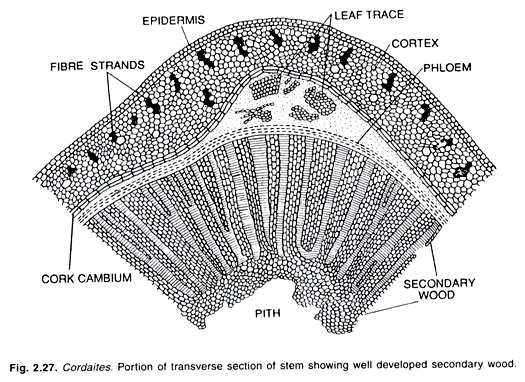
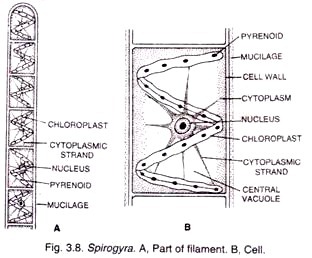
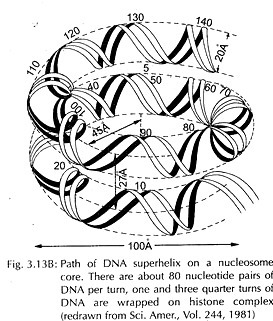
ADVERTISEMENTS:
Read this article to learn about the concept, labelling and translation of gene probes.
Concept of Gene Probes:
The information needed to produce a gene probe may come from many sources but with the development and sophistication of genetic database, gaining this knowledge is usually one of the first stages.
In some cases it is possible to use related proteins from same gene family to gain information on the most useful DNA sequences.
ADVERTISEMENTS:
Protein or DNA sequences that are similar but are from different species may also provide a starting point with which to produce a so called heterologous gene probe.
Also with the help of database, single stranded oligonucleotide probe can be synthesized chemically. This is done by computer controlled gene synthesizers that link dNTPs together on the basis of a desired sequence. Whatever way is used to obtain a probe but it should be assured that probe is unique, and is neither able to self-anneal nor is self-complimentary.
Where scant DNA information is available to prepare a gene probe it is possible in some cases to use the knowledge obtained form analysis of protein as from genetic code it is possible to predict the various DNA sequences which could code for the protein and thus then can synthesize oligonucleotide sequence chemically.
However, due to degeneracy of genetic code there could be more than one oligonucleotide for a given polypeptide. Ideally, sequence that is specific for a given gene and is not longer than 20 bases that contains as many tryptophan and methionine as possible is sufficient as these have unique codons and therefore, fewer possible base sequence that can code for that part of protein.
Labelling of Gene Probes:
An essential feature of gene probe is that it can be visualized by some means. Therefore, labeling of probe is a must and is done by two methods, traditionally by using radioactive labels and by non-radioactive labels. Mostly used radioactive labels are phosphorous 32 (32P), sulphur 35 (35S) and tritium (3H) which are detected by process of autoradiography.
Non-radioactive probes, although less sensitive than radioactive probes but are safe to use. The labelling systems are termed either direct or indirect. Direct labelling allows an enzyme reporter such as alkaline phosphatase to couple directly to DNA. Indirect labelling method which is more popular involves binding of nucleotide that has label attached, e.g., Biotin, fluorescein and digoxigenin.
These molecules are covalently linked to nucleotides. Specific binding proteins may then be used as a bridge between nucleotide and reporter protein such as enzyme. For example biotin incorporated into DNA molecule is recognized with a very high affinity by Streptavidin. This may be in turn is coupled with enzyme alkaline phosphatase which is able to convert colourless compound p-nitro phenol phosphate (PNPP) into yellow colour compound p- nitro-phenol (PNP) and also offers a means of signal amplification.
Thus, rather than the detection system relying on autoradiography, which is necessary for radiolabels, a series of reaction resulting in colour, a light or a chemiluminescence are much better, since autoradiography may take 1-3 days, where colour and chemiluminescence reaction takes a few minutes.
End Labelling of DNA Molecules:
The simplest form of labelling is by 5′- or 3′-end labelling. 5′-end labelling involves a phosphate transfer or exchange reaction where the 5′-phosphate of the DNA to be used is replaced by 32P. This is carried by two enzymes: the first, alkaline phosphatase; removing phosphate group followed by poly nucleotide kinase catalyzing the transfer of phosphate group (32P) to 5′ end of DNA. The newly labelled probe is then purified, usually by chromatography through a sephadex column to remove any unincorporated radiolabel.
Using other end of DNA molecule, the 3′- end is slightly less complex. Here a new, labelled dNTP ([a32-P]ATP or biotin-labelled dNTP) is added to the 3′-end of DNA by enzyme terminal transferase. Although this is a simpler reaction, a potential problem exists because a new nucleotide is added to the existing sequence and so the complete sequence of the DNA is altered, which may affect its hybridization to its target sequence. End-labelling methods also suffer from the fact that only one label is added to DNA so such methods are of low activity in comparison to other who incorporate label throughout the length of sequence of DNA.
Random Primer Labelling:
ADVERTISEMENTS:
The DNA to be labelled is first denatured and is then allowed to re-natured in presence of random sequences of hexamers or hexanucleotide. These hexamers will by chance bind to DNA sample wherever they encounter a complementary sequence and so the DNA will acquire hexanucleotide annealing to it.
Each of the hexamers can act as primer for synthesis of a fresh strand of DNA catalysed by DNA polymerase since it has free 3′- hydroxyl group. The Klenow fragment of DNA polymerase is used for random primer labelling since it lacks 5′ → 3′ exonuclease activity but still acts as 5′ → 3′ polymerase.
Nick Translation:
It is a traditional method of labelling DNA. Low concentrations of DNase I is used to create occasional single strand nick which is then filled in by DNA polymerase using an appropriate dNTP at the same time making a new nick at 3′-side of the previous one. If labelled dNTPs are used, the DNA can be labelled to a very high specific activity.