ADVERTISEMENTS:
The following points highlight the seven steps involved in the preparation of a recombinant DNA. The steps are: 1. Selection of Target DNA 2. Selection of a Suitable Cloning Vector DNA or Vehicle DNA 3. Selection of Restriction Endonucleases 4. Procedure for Production of Recombinant DNA (rDNA) 5. Introduction of the rDNA into a Host Cell 6. Selection of the Transformed/Transfected Cells and 7. Isolation of the Cell Containing Insert-vector rDNA Holding the Gene of Interest
Step # 1. Selection of Target DNA:
Target DNA is selected considering the following points:
(a) It should be easily extractable from its source of natural existence.
ADVERTISEMENTS:
(b) It should be able to be incorporated in the vector at such a place where it can be replicated, transcribed and translated as desired.
(c) The gene product (protein) produced should either be commercially important or important for research purpose.
(d) The foreign DNA (gene) of interest may be viral, bacterial, of plant or animal origin.
The genes for the following proteins are generally cloned i.e. inserted into the vector DNA and the recombinant DNAs produced and the protein extracted for human use.
Step # 2. Selection of a Suitable Cloning Vector DNA or Vehicle DNA:
The cloning vector is the DNA molecule into which the target DNA is introduced producing the recombinant DNA molecule. A good cloning vehicle is one which has only a single site for cutting by a particular restriction endonuclease. There are different types of vectors which can be used to clone fragments of foreign DNA and propagate (clone) them in a suitable host.
Plasmid:
These are extra-chromosomal genetic elements found in a variety of bacterial species. They are double stranded, closed circular DNA molecules that range in size from 1 kb to 200 kb. Often, plasmids contain genes coding for enzymes that under certain circumstances are advantageous to the bacterial host.
The phenotypes conferred by different plasmids are:
i. Resistance to antibiotics.
ii. Production of antibiotics.
iii. Degradation of complex organic compounds.
iv. Production of colicins.
ADVERTISEMENTS:
v. Production of enterotoxins.
vi. Production of restriction and modification enzymes.
To be useful as cloning vector, the plasmid should possess several properties, like:
i. It should carry one or more selectable markers to allow identification of trans-formants and to maintain plasmid in bacterial population.
ADVERTISEMENTS:
ii. It should contain single recognition site for one or more restriction enzymes in the regions of plasmid that are not essential for replication.
iii. It should be relatively small and should replicate in a relaxed fashion.
iv. Preferably these restriction sites, into which foreign DNA can be inserted, should be located within the genes coding for selective markers, so that insertion of a foreign DNA fragment inactivates the gene.
Some commonly used plasmids as vectors for the preparation of recombinant DNA are-pMB-9, pBR- 322, pBR-325, pKC-7, pAC-4, C-184, pAC-105, pMK-16, pMF-3, pBRHl, pUB-110, and pCB-16.
Bacteriophages:
ADVERTISEMENTS:
Bacteriophages are commonly known as phages. They are the viruses that infect bacteria. Similar to other viruses, the phages are very simple containing a protein coat called capsid, enclosing DNA or RNA molecules as the genome. The genes in this genome include the replication gene for DNA/RNA of the phage and the gene for protein coat.
Bacteriophage Lambda (λ):
The infection is caused by the phage particle by attacking to the outside of the bacterium and injecting its DNA chromosome into the cell. This DNA, about 50 kb, linear-double stranded, with single stranded complementary ends of 12 nucleotides in length (cohesive ends), gets arranged in circle in the host cell through pairing of cohesive ends and is transcribed as circular molecule, during early phase of infection. During this phase the phage may either adopt lytic phase or lysogenic phase.
In the course of lytic phase the phage DNA remains independent in the cell, replicates, codes for capsid proteins and forms large number of phage particles within the cell, resulting in the lysis (breakage) of the bacterial cell and thus releasing the phage particle. In the course of lysogenic phase the phage DNA gets incorporated into the bacterial chromosome and exists so for numerous cell divisions, meanwhile synthesizing capsid components and releasing phage particles occasionally. This may convert to lytic phase at any time.
Bacteriophage M13:
ADVERTISEMENTS:
It is a very small, single stranded DNA phage of 10 Kb. The injection of an M13 DNA molecule into an E. coli occurs via the pilus i.e. the structure that connects two cells during sexual conjugation. Inside the cell the DNA synthesizes a complementary strand and becomes the double stranded intermediate known as the replicative form (RF).
The original strand is (+) strand and the complementary strand is the (—) strand made in the bacteria. Only the (+) strand is packaged into the new phage coats. RF of Ml3 acts like a plasmid, which can be easily obtained from E. coli cells and used for rDNA technology. Some of the M13 vectors are M13 mp2, M13 mp5, fdlOl, fdl07, fd-tet.
Cosmids:
Cosmid is a cloning vector consisting of the lambda cos site (single stranded cohesive extensions at the ends of the DNA molecule) inserted into a plasmid, used to clone DNA fragments upto 40 Kb in size. The maximum size of the DNA that can be introduced into any plasmid is 5 kb and that for bacteriophage Ml3 is less than 3 Kb.
Hence it will become difficult to insert and clone large genes, especially those of eukaryotic DNA. Ex. The alpha-2 collagen gene of chicken is 38 Kb. Hence the bacteriophage lambda genome was modified by Collins and John in 1978 by deleting some bases and introducing single stranded complementary extension sequences at the ends of its DNA molecule called the Lambda Cos site, in order to facilitate insertion and cloning of large DNA molecules.
Due to their capacity to hold large sized DNAs, cosmids are used to construct genomic library i.e. a set of recombinant genes that contains the entire DNA present in an individual organism. The construction of libraries in bacteriophage X vectors has proven to be an effective means of isolating segments of DNA from complex eukaryotic genomes.
Cosmids are identified by:
ADVERTISEMENTS:
1. A drug resistance marker and a plasmid origin of replication.
2. One or more unique restriction sites for cloning.
3. A DNA fragment that carries the ligated cohesive ends (cos site) for bacteriophage X.
4. Small size so as to accommodate the eukaryotic DNA fragments of 40-45 kb in length.
Step # 3. Selection of Restriction Endonucleases:
They are endonucleases (enzymes) that cut DNA molecules only at a limited number of specific nucleotide sequences which are un-methylated palindromes. The action of some endonucleases gives DNA fragments with sticky/cohesive ends whereas others give blunt ended DNA fragments. A few restriction endonucleases are shown in the figure.
Step # 4. Procedure for Production of Recombinant DNA (rDNA):
(a) Preparation of Cloning Vector-insert DNA Constructs:
The target DNA is extracted from source organism which can either be a bacterium, fungi, plant or an animal cell DNA, by various analytical methods for which the cells are first broken open to release the contents either by mechanical disruption (grinding frozen material) or by the use of chemicals like lysozyme, EDTA, the detergent-sodium dodecyl sulphate (SDS) etc., either solely or in combination with one or more chemicals.
ADVERTISEMENTS:
Then the DNA is purified from the cell extract for which the extract is treated with proteases and endonucleases and then the proteins precipitated with phenol and chloroform and finally centrifuged. The DNA will be measured in a spectrophotometer at 260 nm, at this wavelength the absorbance (A260) of 1.0 corresponds to 50pg of double-stranded DNA/ml.
This ultraviolet absorbance can also be used to check the purity of a DNA wherein the ratio of DNA absorbance at 260 nm and 280 nm (A260/A280) is 1.8. The ratio less than 1.8 indicates that the preparation is contaminated, either with protein or phenol. Similar procedure is followed to extract the DNA from the vectors. The target DNA is then inserted into the vector DNA, by various procedures.
Two of them are described below:
(i) rDNA formation by the use of restriction endonuclease creating sticky ends:
To join together two duplex DNAs from different species, the two DNAs are separately acted upon by the same restriction endonuclease (BamHI) giving staggered (cohesive/sticky) two stranded cut. Therefore the staggered ends of the two DNAs will be complementary in sequence. Then the two cut DNAs are heated, mixed and cooled, so the sticky ends will base-pair to produce a new kind of recombinant DNA which is joined by DNA ligase.
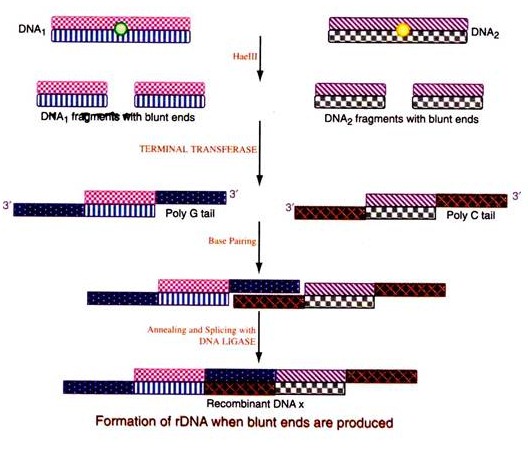
(ii) rDNA formation by use of restriction endonuclease creating blunt ends:
Both the target DNA and the vector DNA are acted Upon by the same restriction endonuclease (Haelll) producing blunt ends. Poly ‘G’ tails are added at the 3′ ends on both strands of the target duplex DNA and poly ‘C’ tails at the 3′ end of the vector DNA by the use of enzyme terminal transferase. Since these two added tails are complementary to each other, they will enable the two DNAs to be paired with the created cohesive ends upon heating and cooling. The nicks are joined by the enzyme ligase.
(b) Preparation of Complementary DNA (cDNA):
It is a cloning technique which involves the conversion of purified mRNA to DNA, prior to its insertion into a vector. Depending upon the source of mRNA its purification procedure varies and in fact many methods for purification of mRNA are available.
The mRNA of interest i.e. beta-globin gene mRNA is obtained by lysing the reticulocytes and the polyribosomes are collected by centrifugation and treated with antibody specific for this protein. The antibody will attach to the just complete protein on the polyribosome and precipitate it. From this precipitate the mRNA is obtained by affinity column chromatography.
The mRNA obtained so, is used as a template and a complementary DNA (cDNA) is made with the enzyme reverse transcriptase. All eukaryotic mRNAs contain poly ‘A’ tails at the 3′ end. Therefore a poly T nucleotide is added, which base pairs with the poly ‘A’ tail of mRNA. This serves as the primer for the enzyme reverse transcriptase that now transcribes the mRNA to make a new complementary strand of cDNA.
The mRNA is then removed and the single stranded cDNA is now replicated by DNA polymerase-I to yield a ‘hairpin’ double stranded DNA. The hair pin and poly A & T tails are cleaved by nucleases. Thus a synthetic double stranded cDNA, specific for the protein beta-globin is produced.
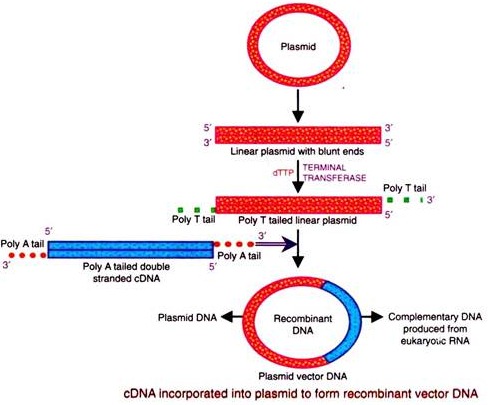
The complimentary DNA prepared above is inserted into a vector (plasmid, phage-DNA etc.). For insertion, a poly ‘A’ tail is added to the opposite 3′ ends of the two strands of the duplex cDNA by terminal transferase. Then the plasmid or vector is opened at a single point to yield the linear DNA by the action of restriction endonuclease producing flush/blunt ends.
Conversely, poly ‘T’ tails are attached to the two 3′ ends of the linear plasmid vector. The tailed linear plasmid and the tailed DNA are allowed to undergo base pairing by heating, mixing and cooling, thereby forming an enlarged circular plasmid containing the new gene i.e. the rDNA. The ends are joined by ligase.
(c) Construction of a DNA Probe:
If the amino acid sequence of a protein/peptide is known then a DNA probe can be chemically synthesized and used to prepare a rDNA and thus clone it in a suitable host. This DNA probe can also be used to screen the gene for this particular protein/peptide in the genomic library.
The DNA probe obtained by this method is not the exact sequence as present naturally in the genomic DNA, because the genetic code is degenerate i.e. more than one codon exists for some of the amino acids. Hence all the possible coding sequences for a given amino acid sequence have to be prepared.
Supposing the following is the amino acid sequence of a part of a peptide:
The nucleotide sequence obtained in any one of the mentioned sequences above, will serve the purpose of formation of rDNA and it’s cloning, resulting in the production of the same desired protein/peptide.
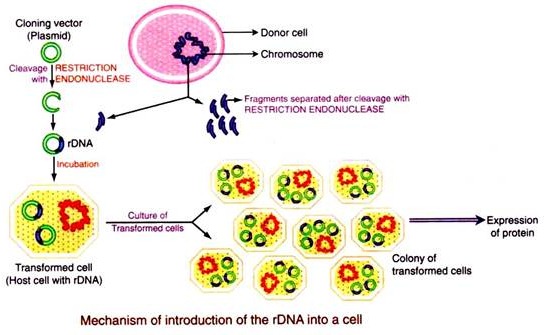
Step # 5. Introduction of the rDNA into a Host Cell:
The rDNA produced in the 4th step above can be cloned (made into many copies) and/or its gene product expressed by introducing it into a suitable host. Various methods are adopted for introduction of the different vectors into different hosts. Any DNA molecule can be introduced into any living cell and the process is called transformation. Transfection is a process of introduction of purified phage DNA.
(a) When the DNA molecule is kept in close proximity of bacterial cells, most species of bacteria are able to take up DNA molecules from the medium without any difficulty.
(b) Some species of bacteria cannot take DNA easily hence they have to be treated physically and/ or chemically in order to make them competent to take up DNA molecules.
(c) E. coli cells incubated with ice-cold solution of 50 mM calcium chloride at 4°C for 30 minutes, become competent to take up DNA molecules, wherein the DNA molecules get attached to the cell exterior. Later the DNA molecules enter the cell upon incubation at 42°C for 2 minutes.
(d) Even after the above mentioned physical/and/or chemical treatment, only 0.01% of the bacterial cells in the same culture gets transformed. Hence the transformed cells have to be selected from non-transformed cells.
(e) Transfection of phage DNA into bacterial cells is also done by the same procedure as that of plasmid introduction into E. coli cells, described above (a) to (d).
(f) Transfection can also be done by packaging the phage DNA into the mature lambda phage particle and then infecting the bacterial cells with it.
(g) The yeast cells of Saccharomyces cerevisiae are transformed by treatment with lithium chloride or lithium acetate.
(h) Calcium phosphate, DEAE dextran and protoplast fusion are used to introduce DNA into the animal cells.
(i) The cell wall of fungal and plant cells are broken by enzymes to get the intact protoplast, which can easily take up DNA. In some cases, the transformation of protoplast is stimulated by a special technique called electroporation. The transformed protoplast reforms the cell wall, divide and regenerate a transformed organism.
(j) An alternative method of introducing any DNA into any of the living cells is by microinjection, using a fine syringe, DNA molecules are directly injected into the nucleus of the cell to be transformed.
Step # 6. Selection of the Transformed/Transfected Cells:
All the cells, incubated in the same culture with the rDNA do not take up the DNA even after physical and/or chemical treatment. Only 0.01% of the total cells incubated becomes competent and takes up DNA to be inserted. Rest of the cells remain without taking up the DNA. Therefore there should be some procedure by which the cells that have taken up the external DNA can be sorted out from those cells which have not taken up the external DNA.
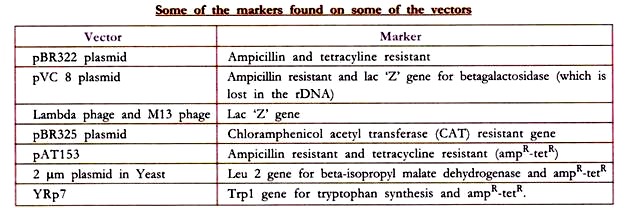
There are various methods for distinguishing the transformed cells from the non-transformed cells, which depends upon the type of vector used and the type of the cell transformed. The basic principle in all these methods is the use of a ‘marker’ i.e. either the vector or the transformed cell will either be resistant or sensitive to an antibiotic and/or will synthesize or be devoid of an enzyme acting on a metabolite and converting it into some recognizable product. Thus the ‘markers’ are the antibiotics and/or enzymes to which the transformed cells respond.
One of the example based on which all the above phenotypic markers are selected, is that of the plasmid cloning vector pBR322. E. coli cells are normally sensitive to the antibiotic ampicillin and tetracycline i.e. they die if the medium in which they are grown is supplied with these antibiotics.
E. coli cells which take up the plasmid vector pBR 322 become resistant to ampicillin and tetracycline antibiotics because pBR 322 has two sets of genes, one set of gene that codes for P-lactamase enzyme that modifies ampicillin into a form that is non-toxic to the bacterium and a different set of genes that codes for enzymes that detoxify tetracycline.
So, those E. coli cells which have taken pBR322 (i.e. transformed cells) can be sorted out from the cells which have not taken pBR322 (i.e. non-transformed cells) by growing all the cells in medium containing ampicillin and tetracycline wherein the cells containing the plasmid pBR322, survive, whereas the cells devoid of pBR322 die.
This is to say that the E. coli cells have transformed from ampicillin and tetracycline sensitive (amps-tets) to ampicillin and tetracycline resistant (ampR-tetR) cells. rDNA prepared using the plasmid pBR322, when entered into an E. coli cell exhibits only ampicillin resistance but not tetracycline resistance, because the restriction endonuclease (Bam-II) used, cleaves pBR322 in the gene for tetracycline resistance, hence this gene gets inactivated.
Step # 7. Isolation of the Cell Containing Insert-vector rDNA Holding the Gene of Interest:
In the 6th step the transformed cells i.e. those cells that have taken up the rDNA are sorted out. Now from these cells the cell containing the gene of interest has to be isolated. The complete human genomic DNA is cleaved into about 7,00,000 pieces by the action of the restriction endonuclease and all of these DNA fragments can be inserted into a cosmid and such an insert containing all the genomic DNA of an organism is called genomic library.
An E. coli genomic library contains all E. coli genes and likewise human genomic library contains all human genes, so on and so forth. From this gene library any one gene, which is commercially important i.e. for example insulin gene, beta-globin gene, growth hormone gene etc., has to be isolated. For this each cell is cloned separately and different procedures are adopted to select the desired clone.
All these procedures are based on two basic themes:
1. Direct Selection of the Desired Gene:
This is just like marker selection procedure, described above, where selection occurs directly in the culture plate itself by the detection of the presence of the protein product of that gene.
2. Identification of the Clone from a Genomic Library:
In this an initial “shot gun” cloning is done to clone all the genes in the genomic library and each gene clone is identified separately by one of the following methods:
(a) By analysing the expression of the gene product.
(b) A DNA probe can be synthesized chemically with a complementary sequence to the desired gene and used to hybridize the DNA from each clone.
(c) A complementary DNA (cDNA) can be prepared from mRNA for a particular gene and then hybridized with the DNA insert.
Hybridization of DNA with another DNA can be done or DNA with RNA or RNA can be hybridized with another RNA, for which different methods are developed.
A few among them are:
i. When DNA-DNA hybridization is made then it is known as southern blotting.
ii. When DNA-RNA hybridization occurs, it is known as northern blot.
iii. When RNA-RNA hybridization occurs, it can be called eastern blot. Actually eastern blot is named for hybridization between protein and ginseng (plant glycoside) so as to identify small molecules like cholesterol, phospholipids etc.
iv. When protein-protein hybridization takes place, it is known as western blotting.
Southern and northern blot hybridization:
DNA is fragmented by restriction endonuclease and then separated by electrophoresis. The DNA is then denatured and filtered on nitrocellulose. To this filter is added radio labelled 32P-DNA probe, with base sequence complementary to the DNA (gene) of interest, which will hybridize (stick/base pair/anneal) to the complementary DNA. The position of hybridization is detected by autoradiography. The radiolabelled probe specific for gene under study can be purified RNA, a cDNA, or a segment of DNA cloned in E. coli.
Western blotting:
In AIDS diagnosis, the component protein of HIV-virus are separated in polyacrylamide gel electrophoresis (PAGE) and trans blotted onto nitrocellulose strips, which are then incubated with individual’s serum. Any HIV antibodies present, bind to viral proteins contained on test strip. The bound antibody visualized by using a conjugate enzyme and chromogenic.
Dot-blot hybridization:
The procedure is the same as southern/northern blotting, but the only difference is that the DNA fragments are not separated by electrophoresis, instead they are directly applied as a dot on nitrocellulose and hence it is known as dot-blot hybridization.
Uses of rDNA technology/gene cloning:
The uses of rDNA technology/gene cloning are enormous and wide spread in all the fields of biological sciences. rDNA technology is being implemented in newer and newer fields of application.
To quote a few uses of rDNA in use at present are:
ADVERTISEMENTS:
(1) By the use of rDNA technology, genes of interest can be inserted, so as to treat various diseases.
(2) Hereditary diseases can be diagnosed in the fetus itself.
(3) rDNA and gene cloning helps in the sequencing of DNA/gene.
(4) Various genes in the chromosomes can be located and the integrated viral genes can be traced out in it.
(5) Mechanism of gene regulation can be studied.
(6) Desired proteins like insulin, hormones, interferon’s, vitamins can be manufactured in bacteria on an industrial basis.
(7) Improved antibiotics can be produced from mixtures of genes from different bacteria.
(8) Recombinant vaccines can be produced by rDNA technology.
Synthesis of human insulin (humulin) using rDNA technology:
Diabetic patients, whose elevated sugar levels are due to impaired insulin production, have been treated with insulin derived from the pancreatic glands of abattoir animals. Though bovine and porcine insulin are similar to human insulin, their composition is slightly different. Consequently, a number of patients’ immune systems produce antibodies against it, neutralizing its actions and resulting in inflammatory responses at injection sites.
This led to synthesizing human insulin i.e. ‘humulin’ by inserting the insulin gene into a suitable vector, the E. coli bacterial cell, to produce insulin that is chemically identical to its naturally produced counterpart. The genetic code for insulin is found in the DNA at the top of the short arm of the eleventh chromosome. It contains 153 nitrogen bases (63 in the chain A and 90 in the chain B).
Manufacturing humulin:
DNA with specific nucleotide sequences for A and B polypeptide chains of insulin are synthesized chemically. 63 nucleotides are required for synthesizing the chain A and 90 for the B chain, plus a codon at the end of each chain, signalling the termination of protein synthesis.
An anticodon, incorporating the amino acid methionine, is then placed at the beginning of each chain which allows the removal of the insulin protein from the bacterial cell’s amino acids. The synthetic A and B chain ‘genes’ are then separately inserted into the gene for a bacterial enzyme β-galactosidase, which is carried in the vector’s plasmid. Insertion of the A gene is carried out by use of the restriction enzyme EcoRl and for B gene Hind-III is used. The recombinant plasmids are then introduced into E. coli cells. The insulin gene is expressed as it replicates with the P-galactosidase as the cell multiplies.
The protein which is formed consists partly of β-galactosidase, joined to both the A and B chains of insulin. The A and B chains are then extracted from the β-galactosidase fragment and purified. The two chains are mixed and reconnected in a reaction that forms the disulfide cross bridges, resulting in pure humulin i.e. synthetic human insulin.