ADVERTISEMENTS:
The work of J. Cairns demonstrated DNA replication in bacteria in an elegant way. He used high resolution autoradiography for observing the distribution of radioactive label in the “chromosomes” of dividing E. coli cells.
The bacterial cells were grown on a medium containing tritiated thymidine. The labelled DNA was extracted from the cells and spread on EM grids. A very thin layer of the photographic emulsion was laid on the grids which were stored in the dark.
Whatever radioactive thymidine was present in the DNA, it reduced the grains of silver halide located directly above it. When the grids were developed and examined in the electron microscope, a track of reduce silver grains (black dots) was observed.
ADVERTISEMENTS:
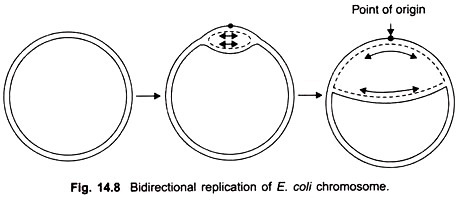
Cairns study revealed that the chromosome of E. coli was a closed circle of duplex DNA; that replication began at a single point of origin and proceeded in a unidirectional manner round the whole circle, until it reached the point of origin.
However, later studies showed that after initiation at the point of origin, replication was bidirectional and proceeds stepwise as follows. First of all an endonuclease enzyme introduces a break or nick in one strand of the duplex at the point of origin thus allowing the two strands to untwist.
The separation of strands is aided by an unwinding protein which binds to a small portion of the nicked strand. The successive binding of additional molecules of this protein leads to opening up of the duplex just before the replication fork. An E. coli cell contains about 10,000 copies of the unwinding protein. The break is perhaps joined by a ligase enzyme.
Replication begins and proceeds in both directions; it is visible in the electron microscope in the form of a replicating “bubble” or “eye” at the point of origin. Both strands of parental DNA are replicated simultaneously (Fig. 14.8), by movement of two replicating forks away from each other.
ADVERTISEMENTS:
A little later the circular replicating DNA appears like the Greek letter theta (θ). The process of unwinding of the circular double helical DNA creates torque which results in supercoiling. An untwisting protein, also called swivelase appears to relieve tension by nicking one strand and allowing some unwinding to take place.
Both strands of the double helix are replicated in short fragments. The DNA polymerase enzymes are not able to polymerise native double stranded DNA. The enzyme DNA dependent RNA polymerase which takes part in transcription of DNA to RNA is able to recognise specific initiation points or chromosomes.
Due to the activity of this enzyme DNA first forms a number of short strands of complementary RNA called primer RNA, each about 100 nucleotides long in the 5′ to 3′ direction. The 3′ end of the primer RNA serves as a starting place for DNA polymerase to add nucleotide units of DNA. The DNA chain is replicated in the 5′ to 3′ direction and is complementary to a single strand of the parent- DNA duplex.
After the DNA chain has attained a length of about 1000 to 2000 nucleotides, an exonuclease enzyme cuts off primer RNA from the 5′ end of DNA. In this way a number of DNA fragments, known as Okazaki fragments are generated during replication instead of one continuous linear strand (Fig. 14.9).
The gaps between the DNA fragments are filled by DNA polymerase I which adds nucleotides complementary to the parental strand in the 5′ to 3′ direction. After the gaps are filled, the remaining nicks are removed by the enzyme ligase, which joins adjacent nucleotides by 3′ 5′ phosphodiester linkages.
In addition to its polymerizing function, DNA polymerase enzyme also performs exonuclease activity by removing bases in 3′ to 5′ direction. If during the synthesis of a DNA chain an incorrect base is added, the DNA polymerase turns back, removes the wrong base in 3′ to 5′ direction, and again starts adding bases in 5′ to 3′ direction.
ADVERTISEMENTS:
The idea that DNA replication takes place by first forming Okazaki fragments received strong support from studies on E. coli mutants deficient in DNA ligase enzyme. In such cells the fragments were seen to accumulate in very large numbers.
Cell division in bacteria is accomplished by replication of the single circular DNA chromosome into two identical daughter duplexes. The replication fork in E. coli moves at the rate of about 20-30 µm per minute. Replication of the entire genome is completed in about 40 minutes. This is followed by partitioning of cell cytoplasm into two daughter cells.
The nuclear membrane is absent; there is no mitosis or spindle formation as in eukaryotes. It seems likely that there is some association between the replicating DNA duplex and the plasma membrane. Electron micrographs of Bacillus subtilis have shown that the plasma membrane invaginates to form a mesosome, and the daughter chromosomes become attached to either side of this invagination.
There is some evidence from in vivo DNA transformation experiments that both the origin and terminal end of replicating DNA are preferentially bound to the membrane. However, as yet, the association between replicating DNA and membrane remains unclear. Further growth of the mesosome partitions the parent cell into two daughter cells.
ADVERTISEMENTS:
Modification of Replicated DNA:
After synthesis the bacterial DNA becomes modified to a conformation characteristic for that bacterium. There is a class of modification enzymes known as DNA methylases which add a methyl group to a newly synthesised adenine (methylate A), or sometimes to a cytosine. There is a specific DNA methylase for each species.