ADVERTISEMENTS:
Structural organization of chloroplast is signified by the presence of double membrane envelope and soluble phase, the stroma, and an internal membrane system, the thylakoids. Both thylakoid and stromal systems are committed for light reaction and carbon dioxide fixation respectively.
Chloroplast attains diversified shapes. Higher plants exhibit lens shaped chloroplasts in their cytosol. The size measures anywhere between 5 and 10 pm long.
Stroma contains soluble enzymes known as rubisco (ribulose bisphosphate carboxylase-oxygenase), accountable for upto 50% of the total leaf proteins. Its molecular weight of 500,000 consists of eight large subunits and eight small subunits and it is credited with one of the most abundant available protein in nature. It executes photosynthesis by accepting carbon dioxide as its substrate and reduces this to carbohydrate status.
ADVERTISEMENTS:
Several members of monocots show marginal deviation in their CO2 fixation process, generally known as C4 plants. The maize, for example, is a C4 plant in which initial carbon dioxide fixation occurs in leaf mesophyll cells containing chloroplasts, which lack rubisco and ultimately devoid of starch.
The enzyme PEP carboxylase (phospho enol pyruvate carboxylase) acts as a major enzyme, catalyses first half of the reaction by forming four carbon oxaloacetate, which is then converted into aspartic acid and malic acid which are exported to bundle sheath cells, where they are decarboxylated and CO2 is refixed by bundle sheath due to the rubisco and operate the Calvin cycle.
In addition to their role in performing photosynthesis and carbon metabolism, chloroplasts are involved in other vital functions such as the synthesis of amino acids and nucleotides, protein synthesis, pigments and hormones.
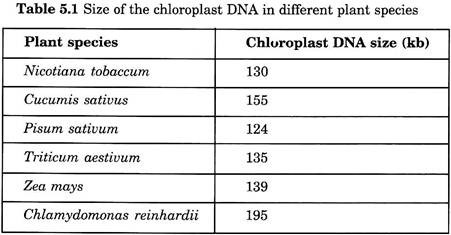
Chloroplast DNA is comparatively large, circular in nature, commonly denoted as ctDNA. The presence of DNA in chloroplast was first identified in 1962. The size of chloroplast DNA is usually 140 kb in higher plants and less than 190 kb in lower eukaryotic plants. However, the size of the ctDNA is generally between 120 and 155 kb.
ADVERTISEMENTS:
By employing DNA-binding flourescent dye several copies of the plastid genome have been visualized. The size of the chloroplast genome can be comparable to bacteriophage T4 (165 kb). There are many copies of circular DNA in chloroplast, i.e., between 20 and 100 copies per chloroplast in higher plants.
In higher plants, chloroplast DNA exists as double-stranded circular molecule. Unlike nuclear DNA, it does not contain 5-methyl cytosine and is not associated with histones. Its buoyant density is around 1.690 gmL-1, which is corresponding to G + C ratio to approximately 37 per cent.
Measurement is based on DNA-DNA association through light on the potential coding capacity of the plastome. The molecular weight of the plastid DNA is between 80 and 100 million, which corresponds between 12,000 and 150,000 base pairs (Table 5.1).
Chloroplast contains one type of chromosome and assumes polyploid status. In young leaves, number of chloroplast attains 200 or more. DNA replication in plastid is semi conservative. In chloroplasts of maize and pea, DNA replication begins at two sites about 7000 base pairs apart and proceeds in both the directions.
Chloroplasts contain introns. They fall into two classes. One of the intron classes is located in tRNA genes and another class in protein coding region. Several photosynthetic related genes that encode proteins are located in thylakoid membrane.
Several evidences confirmed that chloroplast DNA contains 45 genes coding for RNA and 27 genes coding for proteins. These proteins are mainly involved in chloroplast gene expression. The genes coding for proteins of the thylakoid membrane and another 10 gene products are committed for electron transport process.
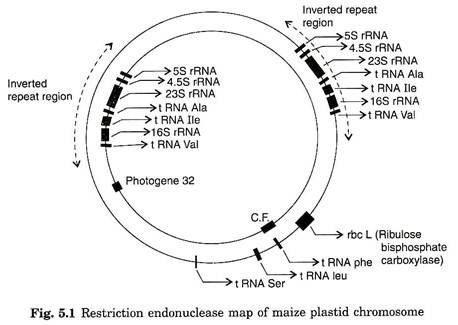
A restriction map for maize chloroplast DNA (139 kb) reveals that plastome contains unique 22,000 base pair inverted repeated sequence, containing the rRNA genes (Fig. 5.1). Some other plastome with similar repeats contains two copies of rRNA genes.
Ribosomes of chloroplast show sedimentation coefficient of 70 S Svedberg units, i.e., 70 S. The ribosomes of cytoplasm exhibits 80 S. The 70 S ribosomes are made up of 23 S and 16 S. Presence of 70 S ribosomes in chloroplast have a resemblance to prokaryotic ribosome, clearly strengthens the hypothesis of its prokaryotic origin.
ADVERTISEMENTS:
Chloroplast ribosomes contain about 50 ribosomal proteins, distributed between the two subunits. The 23 S, 5 S, 4.5 S rRNA are present in the 50 S subunit and the 16 S rRNA is in the 30 S subunit. Plastid contains tRNA synthetase enzymes. The presence of plastid tRNA is able to charge all of the 20 protein amino acids. Synthesis of protein in chloroplast utilizes normal genetic code.
The sequences of the maize and tobacco 16 S rRNA genes are 1491 and 1486 nucleotides in length, respectively. They show 96% sequence homology with each other. Similarly, DNA sequence of 23 S rRNA genes from maize and tobacco is 2898 and 2804 nucleotides respectively.
The distance between 16 S (end) and the 23 S (start) of rRNA gene is 2408 base pairs in maize and 2080 in tobacco (Table 5.2). On the contrary, the distance among prokaryotic organisms is very less, for example, in E. coli distance is 440 base pairs. Longer distance among higher plants is due to the presence of introns upto 950 base pairs.
ADVERTISEMENTS:
During transcription the 16 S, 23 S, 4.5 S rRNA sequence in chloroplast together with the tRNA in the spacer region between 16 S and 23 S genes are transcribed as a polycistronic RNA, which is a precursor RNA undergoes modification to produce mature tRNA and rRNA. Transcription of the rRNA genes takes place at promoter site by chloroplast RNA polymerase upstream from the mature 16 S rRNA sequence and continues till end of the 4.5 S sequences.
Post-transcriptional processing of rRNA such as intron splicing, generation of a number of RNA fragments, the ligation of RNA sequence takes place. Information on the synthesis and processing of chloroplast mRNA is meagre. They seem to be devoid of 5′ cap and do not contain long region of polyadenylic acid at the 3′ end. Some reports suggested that chloroplast mRNA may contain short runs of oligo A.
Plastid Regulatory Sequence:
ADVERTISEMENTS:
Sequencing of plastid genes such as rbcL, rRNA, tRNA, CF polypeptides and photogene 32 have been accomplished, of which rubisco large subunit gene from maize was the first to be sequenced (Mcintosh et al., 1980). There are two putative promoter regulatory sequences (TTGATA and TATGA) present in this region. The putative regulatory sequence rbcL of other species shows deviation (see below).
Expression of rbcL Gene in Chloroplast:
Rubisco gene contains eight subunits of which four are smaller subunits and other four are larger subunits. The genes for larger (L) sububits are coded in chloroplast DNA, and genes for smaller (S) subunits are coded in nuclear DNA. In nuclear code genes have mRNA with 5′ cap and poly-A sequence as evidenced in rubisco gene.
ADVERTISEMENTS:
They are translated on cytosol ribosomes. The transit peptide, which varies from 40 to 60 amino acids in different plants, is transported into chloroplast. After entry of eight smaller subunits inside the chloroplast, signal peptides are cleaved and association between larger subunits and smaller subunits takes place to become functional holoenzyme (Fig. 5.2).
There is a considerable imbalance between the number of nuclear-encoded genes for plastid function and number of plastid-coded genes in photosynthetic cells of higher plants. Several hundreds of gene copies will be produced in chloroplast due to their high copy number; on the other hand, nuclear DNA contains only few copies of the genes for photosynthetic functions. Inspite of this imbalance, some well coordination of gene expression could be seen in the chloroplast of higher plants.