ADVERTISEMENTS:
The following points highlight the seven important enzymes involved in the process of DNA replication of prokaryotes. The enzymes are: 1. DNA Polymerase 2. Primase 3. Polynucleotide Ligase 4. Endonucleases 5. Pilot Proteins 6. Helicase 7. Single-Strand Binding (SSB) Protein.
Enzyme # 1. DNA Polymerase:
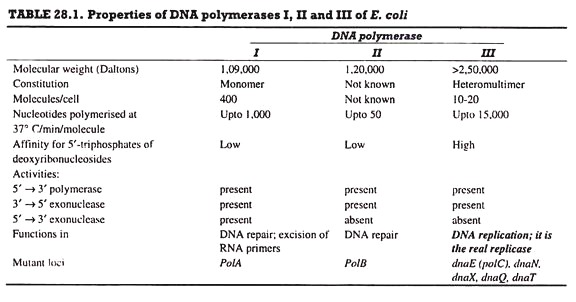
DNA polymerase is the chief enzyme of DNA replication. DNA polymerase activity was discovered by Kornberg in 1956; this activity was due to DNA polymerase I. E. coli has four more enzymes, DNA polymerase II, III (Table. 28.1), IV and V; DNA polymerase III (Pol III) is concerned with DNA replication, while the remaining four enzymes are involved in DNA repair.
All DNA polymerases require the following:
ADVERTISEMENTS:
(1) A template DNA strand,
(2) A short primer (either RNA or DNA), and
(3) A free 3′ -OH in the primer.
They add one nucleotide at a time to the free 3′ -OH of the primer, and extend the primer chain in 5′ → 3′ direction.
ADVERTISEMENTS:
A. DNA Polymerase I:
DNA polymerase I enzyme provides the major part of activity in E. coli. It is chiefly a DNA repair enzyme, and is used for in vitro DNA replication.
This enzyme has the following three activities:
(i) The 5′ → 3′ polymerase activity is responsible for primer extension or DNA synthesis.
(ii) The 5′ → 3′ exonuclease activity is involved in excision of DNA strands during DNA repair; it removes ~ 10 bases at a time. An exonuclease digests nucleic acids (here DNA) from one end, and it does not cut DNA internally.
(iii) The 3′ → 5′ exonuclease activity is responsible for proof-reading.
In this case, only one nucleotide is removed at a time. The polymerase action does commit errors in DNA synthesis. DNA polymerase is known to scrutinize the new bases added to the growing chain and to delete or remove the wrong bases; this is called proof-reading. Proof-reading activity reduces errors in replication by over 100 – fold.
DNA polymerase I is encoded by gene polA, has a single polypeptide, and can initiate replication in vitro at a nick in a DNA duplex. It can be cleaved by proteolytic treatments into a large and a small fragments. This large fragment, called Klenow fragment, lacks 5′ → 3′ exonuclease activity and is used for in vitro DNA replication.
B. DNA Polymerase II:
ADVERTISEMENTS:
DNA polymerase II enzyme functions in DNA-repair. It has 5′ → 3′ polymerase and 3′ → 5′ exonuclease activities, and uses as template only such DNA duplexes that have short gaps.
C. DNA Polymerase III:
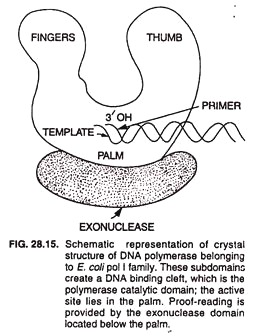
DNA polymerase III enzyme is responsible for DNA replication in vivo. It has 5’→ 3′ polymerase and 3’→ 5′ exonuclease activities. It catalyzes DNA synthesis at very high rates, e.g., 15,000 bases/min at 37°C. It is composed of several subunits. A DNA polymerase molecule has the following 4 functional sites involved in polymerase activity (Fig. 28.15).
(i) Template site binds the strand serving as template during replication.
(ii) Primer site binds to the primer used for DNA replication.
(iii) Primer terminus site binds only to such primers that have free 3′ -OH.
(iv) The nucleotide triphosphate site binds to the deoxynucleotide 5′-triphosphate that is complementary to the corresponding nucleotide of the template. It also catalyzes the formation of phosphodiester bond between the 5′ phosphate of this nucleotide and the 3′ -OH of the terminal primer nucleotide.
ADVERTISEMENTS:
[In addition, the polymerase mole-cule has (5) a 3′ → 5′ exonuclease site and (6) a 5’→ 3′ exonuclease site (in case of DNA polymerase I only)].
In case of eukaryotes, at least nine different DNA polymerases are found; Table 28.2 lists the properties of five of these enzymes. DNA polymerase δ replicates the leading strand, while DNA polymerase ϵ synthesizes the lagging strand.
DNA polymerase α catalyzes priming of both the strands. DNA polymerases ξ, η, τ, and k are all nuclear DNA repair enzymes. DNA polymerase y is found in mitochondria and catalyzes replication of mtDNA.
Enzyme # 2. Primase:
This enzyme activity catalyzes the synthesis of RNA primers to initiate DNA replication. In E. coli, DnaG functions as primase. But in eukaryotes, DNA polymerase α provides this function. There are, however, several other ways in which primers are produced, e.g., the 3′-OH generated by a nick in the template DNA molecule.
Enzyme # 3. Polynucleotide Ligase:
ADVERTISEMENTS:
DNA ligase or polynucleotide ligase catalyzes the formation of phosphodiester linkage between two immediate neighbour nucleotides of a DNA strand. Thus it seals the nicks remaining in a DNA strand either following DNA replication or DNA repair. However, this enzyme cannot fill the gaps in DNA strands.
Enzyme # 4. Endonucleases:
An endonuclease produces an internal cut (single- or double-stranded) in a DNA molecule. But a restriction endonuclease produces cuts only at those sites that have a specific base sequence. During DNA replication, an endonuclease may induce a nick to initiate DNA replication, or it may induce nicks to generate a swivel for DNA unwinding. Restriction endonucleases are required for DNA repair.
Enzyme # 5. Pilot Proteins:
Pilot proteins are produced by most viruses. The type of pilot proteins associated with viral genome determines whether the viral DNA will undergo replication or it would support transcription.
Enzyme # 6. Helicase:
Helicase effects strand separation at the forks and uses one ATP molecule for each base that is separated. In E. coli, DNA functions as helicase; this protein is a hexamer and it moves with the replication fork.
Enzyme # 7. Single-Strand Binding (SSB) Protein:
SSB protein binds to single-stranded DNA, and prevents it from forming duplex DNA or secondary structures. SSB binds as a monomer, but it binds cooperatively in that binding of one SSB molecule facilitates binding of more SSB monomers to the same DNA strand. E. coli SSB is a tetramer.