ADVERTISEMENTS:
Read this article to learn about the techniques and variations of polymerase chain reaction with diagram.
Polymerase Chain Reaction :
The polymerase chain reaction (PCR) is a laboratory (in vitro) technique for generating large quantities of a specified DNA.
Obviously, PCR is a cell-free amplification technique for synthesizing multiple identical copies (billions) of any DMA of interest. Developed in 1984 by Karry Mullis PCR is now considered as a basic tool for the molecular biologist. As is a photocopier a basic requirement in an office, so is the PCR machine in a molecular biology laboratory!
Principle of PCR:
ADVERTISEMENTS:
The double-stranded DNA of interest is denatured to separate into two individual strands. Each strand is then allowed to hybridize with a primer (renaturation). The primer-template duplex is used for DNA synthesis (the enzyme- DNA polymerase). These three steps— denaturation, renaturation and synthesis are repeated again and again to generate multiple forms of target DNA.
Technique of PCR:
The essential requirements for PCR are listed below:
1. A target DNA (100-35,000 bp in length).
2. Two primers (synthetic oligonucleotides of 17-30 nucleotides length) that are complementary to regions flanking the target DNA.
ADVERTISEMENTS:
3. Four deoxyribonucleotides (dATP, dCTP, dCTP, dTTP).
4. A DNA polymerase that can withstand at a temperature upto 95° C (i.e., thermo-stable).
The reaction mixture contains the target DNA, two primers (in excess), a thermo-stable DNA polymerase (isolated from the bacterium Thermus aquaticus (i.e., Taq DNA polymerase) and four deoxyribonucleoties. The actual technique of PCR involves repeated cycles for amplification of target DNA.
Each cycle has three stages:
1. Denaturation:
On raising the temperature to about 95° C for about one minute, the DNA gets denatured and the two strands separate.
2. Renaturation or annealing:
As the temperature of the mixture is slowly cooled to about 55° C, the primers base pair with the complementary regions flanking target DNA strands. This process is called renaturation or annealing. High concentration of primer ensures annealing between each DNA strand and the primer rather than the two strands of DNA.
3. Synthesis:
ADVERTISEMENTS:
The initiation of DNA synthesis occurs at 3′-hydroxyl end of each primer. The primers are extended by joining the bases complementary to DNA strands. The synthetic process in PCR is quite comparable to the DNA replication of the leading strand.
However, the temperature has to be kept optimal as required by the enzyme DNA polymerase. For Taq DNA polymerase, the optimum temperature is around 75° C (for E. coli DNA polymerase, it is around 37° C). The reaction can be stopped by raising the temperature (to about 95° C).
The 3 stages of PCR in relation to temperature and time are depicted in Fig. 8.1. Each cycle of PCR takes about 3-5 minutes. In the normal practice, the PCR is carried out in an automated machine.
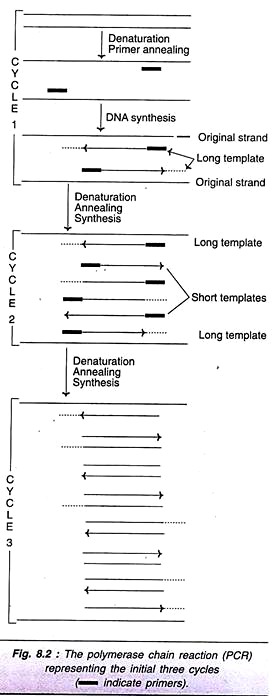
As is evident from the Fig. 8.2 (cycle I), the new DNA strand joined to each primer is beyond the sequence that is complementary to the second primer. These new strands are referred to as long templates and they will be used in the second cycle.
For the second cycle of PCR, the DNA strands (original + newly synthesized long template) are denatured, annealed with primers and subjected to DNA synthesis. At the end of second round, long templates, and short templates (DNA strands with primer sequence at one end, and sequence complementary to the other end primer) are formed.
In the third cycle of PCR, the original DNA strands along with long and short templates are the starting materials. The technique of denaturation, renaturation and synthesis are repeated. This procedure is repeated again and again for each cycle. It is estimated that at the end of 32nd cycle of PCR, about a million-fold target DNA is synthesized (Table 8.1). The short templates possessing precisely the target DNA as double- stranded molecules accumulate.
Sources of DNA Polymerase:
In the original technique of PCR, Klenow fragment of E. coli DNA polymerase was used. This enzyme, gets denatured at higher temperature, therefore, fresh enzyme had to be added for each cycle. A breakthrough occurred with the introduction of Taq DNA polymerase from thermophilic bacterium, Thermus aquaticus. The Taq DNA polymerase is heat resistant; hence it is not necessary to freshly add this enzyme for each cycle of PCR.
Key Factors for Optimal PCR:
ADVERTISEMENTS:
Primers:
Primers play a significant role in determining PCR. The primers (17-30 nucleotides) without secondary structure and without complementarity among themselves are ideal. The complementary primers can hybridize to form primer dimer and get amplified in PCR. This prevents the multiplication of target DNA.
DNA polymerase:
ADVERTISEMENTS:
As already described, Taq DNA polymerase is preferred as it can withstand high temperature. In the hot-start protocol, DNA polymerase is added after the heat denaturation step of the first cycle. This avoids the extension of the mismatched primers that usually occur at low temperature.
Taq polymerase lacks proof reading exonuclease (3′-5′) activity which might contribute to errors in the products of PCR. Some other thermo-stable DNA polymerases with proof-reading activity have been identified e.g., Tma DNA polymerase from Thermotoga maritama; Pfu DNA polymerase from Pyrococcus furiosus.
Target DNA:
In general, the shorter the sequence of target DNA, the better is the efficiency of PCR. However, in recent years, amplification of DNA fragments up to 10 kb has been reported. The sequence of target DNA is also important in PCR. Thus, CC-rich regions of DNA strand hinder PCR.
Promoters and inhibitors:
Addition of proteins such as bovine serum albumin (BSA) enhances PCR by protecting the enzyme DNA polymerase. Humic acids, frequently found in archeological samples of target DNA inhibit PCR.
Variations of PCR:
The basic technique of the PCR has been described. Being a versatile technique, PCR is modified as per the specific demands of the situation. Thus, there are many variations in the original PCR; some of them are discussed, hereunder.
Nested PCR:
ADVERTISEMENTS:
Sequence similarities between the target DNA and related DNA are very frequently seen. As a result of this, the primers may bind to both the DNAs and therefore even the undesired DNA also gets amplified in PCR. Use of nested primers increases the specificity of PCR, and selectively amplifies target DNA. Nested PCR is illustrated in Fig. 8.3. In the first cycle of PCR, the products are both from target DNA and undesired DNA. A second set of internal primers is now used. They will selectively bind to target DNA and amplification proceeds.
Inverse PCR:
In the inverse PCR, amplification of DNA of the unknown sequences is carried out from the known sequence (Fig. 8.4). The target DNA is cleaved with a restriction endonuclease which does not cut the known sequence but cuts the unknown sequence on either side. The DNA fragments so formed are inverted and get circularized (DNA ligase is employed as a sealing agent).
The circle containing the known sequences is now cut with another restriction enzyme. This cleaves only the known sequence. The target DNA so formed contains the known sequence at both the ends with target DNA at the middle. The PCR amplification can now be carried out. It may be noted that the primers are generated in the opposite direction to the normal, since the original sequence is inverted during circularization.
Anchored PCR:
ADVERTISEMENTS:
In the anchored PCR, a small sequence of nucleotides can be attached (tagged) to the target DNA i.e., the DNA is anchored. This is particularly useful when the sequence surrounding the target DNA is not known. The anchor is frequently a poly G tail to which a poly C primer is used. The anchoring can also be done by the use of adaptors. As the adaptors possess a known sequence, the primer can be chosen.
Reverse Transcription PCR:
The PCR technique can also be employed for the amplification of RNA molecules in which case it is referred to as reverse transcription — PCR (RT-PCR). For this purpose, the RNA molecule (mRNA) must be first converted to complementary DNA (cDNA) by the enzyme reverse transcriptase. The cDNA then serves as the template for PCR. Different primers can be employed for the synthesis of first strand of cDNA. These include the use of random primers, oligo dT primer and a sequence specific primer (Fig. 8.5).
Asymmetric PCR:
PCR technique can also be used for the synthesis of single-stranded DNA molecules, particularly useful for DNA sequencing. In the asymmetric PCR, two primers in a ratio of 100: 1 are used. After 20-25 cycles of PCR, one primer is exhausted. The result is that in the next 5-10 PCR cycles, only single-stranded DNAs are generated.
Real-Time Quantitative PCR:
The quantification of PCR products in different cycles is not as simple as projected by theoretical considerations (Table 8.1). In practice, large variations occur. The most commonly used technique for measuring the quantity of PCR is by employing a fluorescence compound like eithidium bromide.
The principle is that the double-stranded DNA molecules bind to ethidium bromide which emit fluorescence that can be detected, and DNA quantified. The synthesis of genes by PCR and the role of PCR in site-directed mutagenesis are described elsewhere.
Random Amplified Polymorphic DNA (RAPD):
Normally, the objective of PCR is to generate defined fragments of DNA from highly specific primers. In the case of RAPD (pronounced as rapid), short oligonucleotide primers are arbitrarily selected to amplify a set of DNA fragments randomly distributed throughout the genome. This technique, random amplified polymorphic DNA is also known as arbitrarily primed PCR (AP-PCR).
The procedure of RAPD is comparable to the general technique of PCR. This method basically involves the use of a single primer at low stringency. A single short oligonucleotide (usually a 9-10 base primer) binds to many sites in the genome and the DNA fragments are amplified from them. The stringency of primer binding can be increased after a few PCR cycles. This allows the amplification of best mismatches.
RAPD can be carefully designed so that it finally yields genome- specific band patterns that are useful for comparative analysis. This is possible since genomic DNA from two different individuals often produces different amplified patterns by RAPD. Thus, a particular DNA fragment may be generated for one individual and not for the other, and this represents DNA polymorphism which can be used as a genetic marker.
RAPD is widely used by plant molecular biologists for the genetic identification of plant species. For this purpose, different combinations of nucleotides, most of them random oligonucleotide primers have been designed and are commercially available. As each random primer anneals to a different region of DNA, many different regions of loci on the DNA can be identified. RAPD is thus useful for the construction of genetic maps and as a method for genomic fingerprinting.
Limitations of RAPD:
The main problem of RAPD is associated with reproducibility. It is often difficult to obtain similar levels of primer binding in different experiments. It is therefore difficult to correlate results obtained by different research groups on RAPD.
Amplified Fragment Length Polymorphism (AFLP):
AFLP is a very Sensitive method for detecting polymorphism in the genome. It is based on the principle of restriction fragment length polymorphism and RAPD. AFLP may be appropriately regarded as a diagnostic fingerprinting technique that detects genomic restriction fragments.
In the AFLP, PCR amplification rather than Southern blotting (mostly used in RFLP) is used for the detection of restriction fragments. It may be noted that AFLP is employed to detect the presence or absence of restriction fragments, and not the lengths of these fragments. This is the major difference between AFLP and RFLP. AFLP is very widely used in plant genetics.
It has not proved useful in the mapping of animal genomes, since this technique is mainly based on the presence of high rates of substitutional variations which are not found in animals. On the other hand, substitutional variations resulting in RFLPs are more common in plants. The basic principle of AFLP involves the amplification of subsets of RFLPs using PCR (Fig. 8.6).
A genomic DNA is isolated and digested simultaneously with two different restriction endonucleases — EcoRI with a 6 base pair recognition site and Msel with a 4 base pair recognition site. These two enzymes can cleave the DNA and result in small fragments (< 1 kb) which can be amplified by PCR. For this purpose the DNA fragments are ligated with EcoRI and Msel adaptors.
These common adaptor sequences (flanking genomic sequences) serve as primer binding sites on the restriction fragments. The DNA fragments can be amplified with AFLP primers each having only one selective nucleotide. These PCR products are diluted and used as templates for the selective amplification employing two new AFLP primers that have 2 or 3 selective nucleotides.
After the selective amplification by PCR, the DNA products are separated on a gel. The resultant DNA fingerprint is identified by autoradiography. AFLP fragments represent unique positions in the genomes, and hence can be used as landmarks to bridge the gaps between genetic and physical maps of genomes. In plants, AFLP is useful to generate high density maps, and to detect genomic clones.
Rapid Amplification of cDNA Ends (RACE):
As already described (See p. 115), reverse transcription, followed by PCR (RT-PCR) results in the amplification of RNA sequences in cDNA form. But the major limitation of RT-PCR is related to incomplete DNA sequences in cDNA. This problem is solved by using the technique rapid amplification of cDNA ends. RACE is depicted in Fig. 8.7, and briefly described below.
The target RNA is converted into a partial cDNA by extension of a DNA primer. This DNA primer was first annealed at an interval position of RNA, not too far from the 5′-end of the molecule. Now addition dATP (As) and terminal deoxynucleotidyl transferase extends the 3′-end of the cDNA.
This happens due to the addition of a series of as to the cDNA. These as series now act as the primer to anneal to the anchor primer. A second strand of DNA can be formed by extending the anchor primer. The double-stranded DNA is now ready for amplification by PCR. The above procedure described is called 5′- RACE, since it is carried out by amplification of the 5′-end of the starting RNA. Similar protocol can be used to carry out 3′-RACE when the 3′-end RNA sequence is desired.
Limitations of RACE:
Since a specific primer is used, the specificity of amplification of RACE may not be very high. Another disadvantage is that the reverse transcriptase may not fully reach the 5′-ends of RNA, and this limits the utility of RACE. In recent years, some modifications have been done to improve RACE.