ADVERTISEMENTS:
The below mentioned article provides an overview on Ribonucleic Acid (RNA).
In broad features, the chemistry of RNA is comparable to the chemistry of DNA.
The chemical analysis shows the following two main differences in the polynucleotide chains of DNA and RNA:
ADVERTISEMENTS:
(i) The sugar in RNA is ribose whereas it is deoxyribose in DNA.
(ii) While in DNA, the common bases are adenine, guanine, thymine and cytosine, the RNA contains adenine, guanine, cytosine and uracil (i.e., in RNA molecule, uracil takes the place of thymine). Chemical analysis of RNA molecules obtained from different sources has revealed that base pair ratios (i.e., A/U = C/G) are not equal in those molecules.
RNA is found in the form of a polynucleotide chain which consists of 4 monomeric ribotide or ribonucleotides, namely, AMP, GMP, CMP and uredine monophosphate or UMP.
DNA acts as a template for RNA synthesis. The RNA molecule is assembled from four ribonucleoside 5-triphosphates namely adenosine triphosphate (ATP), guanosine triphosphate (GTP), Cytidine triphosphate (CTP) and uridine triphosphate (UTP) on the surface of one of the two strands of DNA in presence of a specific enzyme called RNA Polymerase. This process is called transcription (Fig. 10.2 A).
ADVERTISEMENTS:
The reaction is given below:
The primary reaction during the RNA chain growth is also comparable, since the two terminal phosphates of ribonucleoside triphosphate are split off while a nucleotide unit with one phosphate group is added to the 3′ end of the chain.
Since ribose and deoxyribose differ only in one oxygen atom, while uracil and thymine differ only in a methyl group, inter-conversion of RNA and DNA precursors takes place very readily in living cells (Du Praw, 1970). Single strands of RNA are polarized 3′-5′ polyribotide chains and complementary RNA chains also pair into double helices very readily and hydrogen bonds are established between G—C and A—U base pairs.
The three-dimensional structure of RNA is less defined. RNA is commonly found in single stranded state. It does have regions of double helical configuration where the same chain loops upon itself forming hydrogen bonded base pairing structure which may sometime be folded on itself to form localized helical regions.
The relationship of RNA polymerase to its template DNA is different from that of DNA polymerase reaction in the following two respects:
(i) There is a notable preference for a helical DNA as template and as reported by Hurwitz et al. (1963), the two strands of DNA are not dispersed during the reaction.
(ii) Only one of the two complementary strands of DNA is copied (or transcribed) to determine the sequence of ribotides in the RNA chain (Hayashi et al., 1964).
The rarity of complementary RNA strands follows from the mechanism of RNA synthesis which produces only one chain at a time.
ADVERTISEMENTS:
Transcription:
Genome contains complete information for the structure and function of organism, but not all genes are active at any one time. DNA does not act as template for the synthesis of proteins. The genetic information of DNA is transferred to RNA molecules. This process is called transcription. It is actually RNA that acts as a template to produce sequence of aminoacids or protein. This process is called translation.
The common features of transcription in both prokaryotes and eukaryotes are as follows:
1. For transcription two strands of DNA unwind locally and for each gene—only one strand of the double helix DNA acts as template for RNA synthesis. That is called sense strand and the opposite strand of DNA is termed antisense strand.
ADVERTISEMENTS:
2. The product of transcription is single stranded RNA.
3. Ribonucleoside triphosphates-ATP, GTP, CTP and UTP are RNA precursors that are polymerised into RNA chain. Polymerization is catalysed by the enzyme RNA polymerase and Mg++.
4. RNA polymerisation reaction is similar to the of DNA polymerization. RNA polymerase selects ribonucleoside phosphate to form complementary base pairs with the exposed nucleotides of sense strand of DNA.
5. RNA is synthesized in 5′ – 3′ direction e.g., if DNA template strand is3′-TCCGATTT-5′; the RNA chain would be5′-AGGCUAAA-3′.
ADVERTISEMENTS:
The process of transcription on prokaryotic DNA proceeds in the following sequence:
1. Prokaryotic organisms have their own RNA polymerases. Bacteriophages use host’s RNA polymerases or code for their own. Transcription requires double stranded DNA, all four ribonucleoside triphosphates, RNA polymerase enzyme with sigma factor, Mg++ ions and nus A protein factor. RNA polymerases core enzyme is composed of 4 subunits β’, β, and 2 α subunits (Fig. 10.22).
β’ subunit of core enzyme is DNA binding subunit. It binds to the sense strand of DNA at a specific site called promoter. The other DNA strand which is not transcribed is called anti sense (Fig. 10.23).
Each gene (a specific length of DNA on one strand of helix) is divided into coding and non-coding regions (Fig. 10.24). A coding region carries a nucleotide sequence with hereditary information. Coding sequence is flanked on either sides by non-coding sequences.
Non-coding sequence upstream is organised into promoter region with a set of seven nucleotides in the middle, called Pribnow Box. Between promoter and coding sequences is a short stretch of nucleotides called initiator site which signals transcription of the coding sequence.
The non-coding region downstream has a set of nucleotides called terminator which terminates transcription. Thus Promoter allows binding of the enzymes for transcription, the initiator gives the start signal for transcription of coding region and the terminator acts as a stop signal for transcription.
2. About 35 base pairs upstream from the start of RNA synthesis is a recognition site. RNA polymerase probably first binds there and then slides to the Pribnow box to initiate transcription β’ subunit of core enzyme binds to the sense strand of DNA at promoter sequence.
Before binding to DNA, the core enzyme becomes associated with σ (sigma) factor which plays an important role in transcription, a factor is a 95,000 dalton polypeptide. It initiates RNA synthesis at promoter site along the DNA.
ADVERTISEMENTS:
If o factor is not linked to RNA polymerase, the enzyme can initiate RNA synthesis, but at random and then both strands are copied instead of one.
3. RNA polymerase then catalyses denaturation or separation of two DNA strands exposing the template strand for initiation of RNA synthesis (Fig. 10.25).
4. RNA synthesis starts, on DNA template in 5′ to 3′ direction. The ribonucleotides come one after another in an array and form complementary base pairs with the bases of exposed template strand of DNA. The consecutive ribonucleotides are liked by formation of phosphodiester bonds in the same way as in DNA synthesis.
This process is catalysed by P subunit of RNA polymerase. When the polymerisation has started, o factor which acted like a catalyst in initiation of RNA synthesis at specific site dissociates from core enzyme and becomes available for initiating on other transcription site.
5. When the gene has been transcribed, a protein factor called nus-A becomes linked to RNA polymerase and the process of transcription is stopped. Then nus-A, core enzyme or RNA polymerase and RNA strand are separated. The release of RNA strand from DNA template requires the activity of a P (rho) protein.
ADVERTISEMENTS:
6. When newly synthesised RNA is displaced, DNA strands renaturate to form double-helical DNA.
Transcription in Eukaryotes:
Although RNA synthesis in eukaryotes takes place more or less in the same way as in prokaryotes, yet some differences are there:
(i) In controlling sequences, and
(ii) Involvement of more than one RNA polymerase enzyme in eukaryotic organisms. In eukaryotes, three RNA polymerases; RNA polymerase I, RNA polymerase II and RNA polymerase III are known which play special roles in RNA synthesis. All the three RNA polymerases are complex polypeptides, each consisting of several subunits.
As regards transcription controlling sequences, the base pair sequences upstream from coding sequence indicate the presence of consensus sequence TATAAA called TATA box or the Goldberg- Hogness box about 30 basepairs upstream from the coding sequence. That is considered to be promotor sequence. Further upstream from TATA Box, some other sequences also play role in regulating transcription.
Several types of promoter sequences are used in eukaryotes. The terminator sequences downstream of coding sequence are not properly understood in eukaryotes. In yeast, part of termination sequence is TTTTTATA.
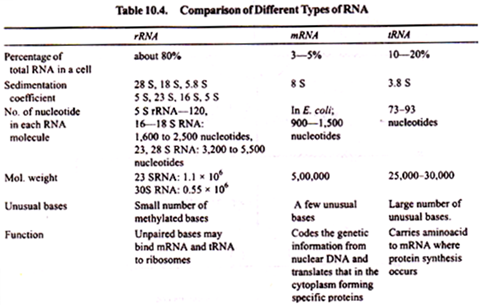
There are different types of RNA which perform different functions. These RNAs are categorized as follows:
A. genetic RNA or Viral RNA
B. non-genetic RNA.
Genetic RNA:
In most of the plant viruses and in some of bacteriophages and animal viruses, the hereditary material is single stranded RNA. In a few cases such as the poliovirus, influenza virus and reovirus, the RNA is double stranded.
The viral RNA is also called genetic RNA because it contains all the information which are normally found in DNA in higher organisms. Molecular weights of RNA in viruses can vary greatly. Larger virus particle has got larger RNA molecule than the smaller ones.
Each Tobacco Mosaic Virus (TMV) with particle weight 4×107 contains a single strand of RNA of molecular weight 2.2 X 106. TMV RNA is one of the most intensively studied and one of the best understood molecules of RNA. The double stranded RNA of viruses follows the same basic rules of base pairing as in DNA.
The genetic RNAs found in viruses act as informational molecules. However, the information is not transferred from RNA to RNA. Here the viral RNA acts as template for DNA synthesis by reverse transcriptase (Reverse transcription). The DNA strand synthesized on viral RNA is involved in the synthesis of complementary viral RNA. Viral RNA cannot replicate by itself while present in the host cell.
The viral RNA functions directly as a messenger RNA which in association with ribosomal apparatus of the host cell directs on one hand the synthesis of proteins that form the viral sheath and on the other hand RNA polymerase enzyme which is required in RNA replication.
With the help of RNA polymerase and base pairing rules, the viral RNA synthesizes a complementary RNA chain on its surface and thus double stranded structure is produced.
Non-Genetic RNA:
The hereditary material in cellular forms of organisms is DNA. RNA, though present in good quantity, does not act as genetic material. It plays very important roles in the synthesis of cellular proteins.
The different types of non-genetic RNA found in the cell are as follows:
(i) Ribosomal RNA (rRNA) which is sedimented at high speed in the ultra-centrifuge.
(ii) Transfer RNA (tRNA) (formerly called soluble RNA or s RNA) which remains in the supernatant after the sedimentation of ribosome.
(iii) Messenger RNA (mRNA) which incorporates labelled uracil before other RNAs.
Ribosomal RNA (rRNA):
It is the most stable RNA which occurs in ribosomes. About 80% of the total cellular RNA occurs in ribosomes. Ribosomal RNA makes up about 40-60% of the total weight of ribosomes. Each ribosome consists of one large and one small subunit. Both subunits must associate to function in protein synthesis. The association and dissociation of two subunits depend on the Mg++ concentration.
The ribosomes are usually characterized by their sedimentation coefficients obtained by analytical ultracentrifugation experiments. The ribosomes of eukaryotes sediment at about 80S whereas in bacteria they sediment at 70S. Both these types of ribosomes are structurally similar and are complexes of rRNA and proteins.
The ribosomes of eukaryotic cells contain; 28S RNA, 5.8S RNA and 5S RNA in the larger (60S) subunit and 18S rRNA in the smaller (40S) subunit. The ribosomes found in prokaryotes and those found in the plastids and mitochondria of eukaryotes also contain three kinds of rRNA; 23S RNA and 5S RNA in larger (5OS) subunit and 16S RNA in smaller (3OS) subunit.
Ribosomal RNA contains the four major RNA bases with slight degree of methylation.
The G/C and A/U ratios in the ribosomal RNAs of different sources vary considerably. The one regularity that has been observed is that rRNA from all sources has a G + C contents more than 50%.
Its molecule appears as single polynucleotide strand which is unbranched and flexible (primary structure) but at low ionic strength, it shows random coiling. At high ionic strength, the molecule shows helical regions with complementary base pairing and looped outer region (Secondary structure) (Fig. 10.26).
Physical studies of rRNA, such as hyperchromicity, melting point and viscosity studies, suggest that a great deal of base pairing occurs within the isolated rRNA molecules, but this is not clear how regularly periodic it is or whether it has any relevance to the structure of the RNA in the ribosomes.
In eukaryotic cells of plants and animals the different types of rRNA molecules are transcribed on rDNA in the nucleolar organizer region of a particular chromosome to which nucleolus is attached.
The important steps in the synthesis and maturation of eukaryotic ribosome are outlined below:
(a) The ribosomal DNA (rDNA) located in the core of the nucleolus synthesizes 45S RNA which is a large initial precursor molecule for 28 S RNA and 18 S RNA.
(b) 45 S RNA is combined with proteins to form a complex of S = 80. The proteins of this complex may include ribosomal and other types.
(c) 45 S RNA is cleaved into 18 S and 32 S pieces. During this period changes in protein composition may take place and side by side loss of some accessory proteins and alterations in the configuration of the two particles may also take place.
(d) 5 S RNA of non-nucleolar origin is incorporated with the 32 S RNA to form a large 65 S subunit which is in due course of time converted into a 60 S or larger subunit of ribosome.
(e) 18 S RNA piece produces 40 S or smaller subunit of ribosome.
(f) The ribosomal subunits along with residual accessory proteins leave the nucleoplasm and come to cytoplasm through annuli of nuclear envelope. In the cytoplasm the two ribosomal subunits (60 S and 40 S) become associated to form a whole ribosome (Fig. 10.27).
As regards the biosynthesis of prokaryotic ribosomes, enough information’s are available about their protein and RNA components.
According to Smith et al. (1968), genes coding for 5 S, 23 S and 16 S rRNAs found in 70S ribosomes of bacteria and according to M Nomura and Associates (1969) 16 S, 23 S and 5 S RNA sequences found in 50 S ribosomes of Escherichia coli are tightly clustered in the region of bacterial DNA and are present in several copies.
The primary transcript of each repeat unit in a precursor-z-RNA (Pre rRNA) of 30 S (p 30 S) which is cleaved to produce p 16 S, p 23 S, and p5 S RNAs, which are immediate precursors of mature 16 S, 23 S and 5S rRNAs respectively. Normally 30 S Pre rRNA molecule is not observed in wild type cells because the cleavaging occurs while p 30 S is still in the process of transcription. (Fig. 10.28).
The pre-rRNAs undergo both tailoring and chemical modification before they become mature rRNAs. During tailoring of larger rRNA molecule, 16 S rRNA sequence is first cleared off and separated from 23 S and 5 S sequences. The fragment containing 16 S information is still larger than the mature 16S rRNA by at least 100 bases and is not methylated.
Methylation and tailoring of this molecule occur after it has associated with a number of proteins to form the precursor ribosomal subunits. 5 S rRNA does not undergo tailoring and methylation before it becomes mature. The whole process of biogenesis of 70 S ribosome occurs in cytoplasm.
The methylation thus occurs after transcription and most of the methyl groups are added to the bases, with only a few added to the 2-OH of ribose moity. .All the methylations of 23 S rRNA occur on the p 30 S RNA component whereas most, if not all, of the methyl groups of 16 S rRNA are added when it is in a mature form.
Thus methylation of 23 S rRNA is an early event and methylation of 16 S rRNA is a late event in the rRNAs processing.
The functions of rRNA are still not properly understood. The main function of rRNA is evidently to serve as the site for finding of single stranded mRNA.
Transfer RNA:
When a cell homogenate containing 10 M of Mg is centrifuged at high speed (100,000 Xg for 120 minutes) the RNAs of high molecular weights bound to ribosomes are sedimented and the supernatant liquid so obtained contains a fraction of RNA which is called transfer RNA or tRNA. Earlier, it was called soluble RNA (sRNA).
RNAs available in the supernatant liquid have a “transfer function” and act as “adaptors” (or connectors) between the amino acids and messenger RNA. Transfer RNAs make up about 15% of the total cellular RNA. Transfer RNAs are the small natural nucleic acids with sedimentation coefficient of 4.
The nucleotide chain length is remarkably uniform for all the tRNAs of prokaryotes and eukaryotes, ranging from 76 to 85 nucleotides. The cells probably contain more than 60 different types of tRNA molecules, molecular weight ranging from 25,000 to 30,000 daltons (one dalton = Atomic weight of hydrogen).
Each molecule has a specificity for a given amino acid to which it can attach by a covalent bond. Holley et al. (1965) showed that tRNA which is specific for alanine in yeast cells consists of 77 nucleotides (mol. wt = 26,600) including 8 A, 12U, 25G, 23C and 9 unsaturated bases (mostly methylated bases).
Medison et al. (1967) have shown that tyrosine tRNA consists of 78 nucleotides and Zachan et al. (1967) have shown that serine tRNA is composed of 85 nucleotides.
The A/U and G/C ratios approach equality, so that there is a possibility for considerable base pairing within the molecule. tRNA is actually a single polynucleotide chain which is apparently bent in the middle and the two arms form loops. Thus the molecule becomes clever leaf like. All tRNA molecules have the sequence CCA at the 3′ end and G residue at 5 end.
(5′)G………… tRNA…………… CCA(3′)
It is at the 3′ end of the chain (CCA-OH end) that the activated amino acid is attached during the protein synthesis. tRNA molecule appears as a single strand of polynucleotides (primary structure). Holley et al. (1965) first proposed the clover leaf pattern of tRNA molecule (Secondary structure) (Fig. 10.29).
Lake and Beeman (1967) have shown that tRNA molecule consists of 3 folds giving it a shape of the clover leaf According to clover leaf model the single polynucleotide chain of tRNA is folded upon itself to form 5 arms. As a result of folding 5′ and 3′ ends of the polynucleotide chain come near each other.
Some of the arms may be differentiated into stem and a loop. In the stem region juxtaposed bases are complementary and paired but in loop there is no pairing.
As regards the structure and function of tRNA molecule the following general conclusions have been made (Fig. 10.30):
(i) The sequence of bases is such that in each case the molecule can form a hydrogen bonded “clover leaf configuration in which CCA bases at 3′ end stick out. The last residue, adenylic acid (A), is the amino acid attachment site. At the 3′ terminal, the sequence CCA — OH is added to all tRNAs post transcriptionally.
(ii) All tRNAs have one part (right loop of the chain) which is common and which consists of seven unpaired bases including pseudouridine (Ѱ). It is known as the TѰC arm. It may be involved in the binding of the tRNA to the ribosome.
(iii) All the tRNA molecules contain 7-15 unusual bases, many of which are methylated or demethylated derivatives of A,U, G and C. Unusual bases are nucleotides of methylinosine (Mel), pseudouridines (Ѱ U or UH2), methylated purines (methylated A and G), methylated pyrimidines such as methylated thyamine or ribothymine, 5 methyl cytosine and others. Not all these unusual bases are present in one tRNA.
(iv) There is one nucleotide triplet in the bottom loop of the chain which is different in all tRNAs examined so far. That is called anticodon. The anticodon is site which probably is commentary to the codon on the messenger RNA and is the site of base pairing between tRNA and mRNA. The nucleotide to the 5′ side of anticodon is always U and that to the 3′ side is always a modified purine.
(v) In the left loop of all tRNA there occurs DHU (Dihydrouridine) base. The DHU arm has a site for the recognition of the amino acid activating enzymes.
(vi) One of the most important structural similarities among the clover patterns of various tRNA molecules is that the overall distance from CCA at one end to the anticodon at the other end appears to be constant (Fig. 10.31).
The difference in nucleotide numbers in different molecules being compensated for by the size of the little loop or “extra arm” located between the right loop and the bottom limb (Figs. 10.29 and 10.30). Unusual bases, such as, dimethyl guanosine, dime A, MeG are located in regions not forming hydrogen bonds. Dimethyl guanosine usually occurs in the comer between the left and bottom loops.
This suggests that the role of usual bases is to help to determine the three-dimensional structure of tRNA which is of crucial importance. Tertiary structure of tRNAs. The application of x-ray crystallography to the stable crystals of tRNA has revealed the tertiary structure of tRNA including the location of the major groups.
The data revealed that all the double helical stems predicted by the clover leaf model do exist and, in addition, some hydrogen bonds bend the clover leaf model into a stable, rough L-shaped appearance, in which the amino acid binding CCA-OH group at 3′ end of the chain is located at the opposite end from the anticodon loop.
Biosynthesis of tRNA:
Unlike mRNAs the tRNA molecules are modified extensively. In-deed, mature tRNAs not only have modified bases but are considerably shorter than the primary gene transcript. In both prokaryotes and eukaryotes primary gene transcripts called Precursor-tRNAs (Pre-tRNAs) are produced which are later clipped and trimmed to produce mature tRNA molecule.
In prokaryotes pre-tRNAs contain either single tRNA molecule with extra leader and trailor sequences at the 5′ and 3′ ends or contain several species of tRNA molecules each. In the latter case the tRNA molecules are separated by spacer nucleotide sequences.
At least two enzymes take part in the processing of pre-tRNAs:
(i) Ribonuclease P catalysing the removal of leader sequence at 5′ end and
(ii) Ribonuclease Q catalysing the removal of 3′ trailor sequence. Another enzyme is also involved in the removal of spacer sequences from multi tRNA precursor to liberate tRNA molecules.
In eukaryotes the pre-tRNAs are larger by some 15-35 nucleotides than die mature tRNAs. The processing of pre-tRNAs presumably occurs, although, the enzymes involved and the sites of their action are not yet known.
The transfer RNAs show specificity in attaching to amino acids (Fig. 10.32). So, there must be at least 20 different tRNAs in the cytoplasm of a cell.
The function of tRNA is to carry a particular amino acid at a definite place on the mRNA.
Four recognition sites in tRNA to achieve this purpose are as follows (Fig. 10.32):
(i) The amino acid attachment site which is CCA sequence at the 3′ terminal.
(ii) Antlcodon which is a group of middle three bases on the bottom loop of tRNA molecule. Anticodon of tRNA recognises three complementary bases of a codon in mRNA molecule.
(iii) Ribosome recognition site which is common to all tRNA molecules and is found in TѰC loop. This consists of GTѰCG sequence.
(iv) Amino acid activating enzyme recognition site or syntheses site found in left arm which activates the enzyme and charges specific amino acid with tRNA.
Messenger RNA:
Messenger RNA. Jacques Monod and Francois Jacob (1961) of Pasteur Institute in Paris coined the term “messenger RNA” to describe the template RNA that carried genetic information for protein synthesis from nuclear DNA to ribosomes in the cytoplasm, Messenger RNA makes up less than 5% of the total cellular RNA.
Messenger RNA is normally attached to the smaller subunits of Ribosomes and the length of mRNA is such that it connects a number of ribosomes and, indeed, such clusters of ribosomes called polyribosomes or polysomes have been observed with electron microscopes. mRNA is assembled on a DNA template.
Since DNAs of different organisms differ only in the sequence of their bases, mRNAs which are formed from them must reflect different base sequences. The mRNA is complementary copy of DNA. This was demonstrated experimentally in 1960 by Marmur and Lane.
They showed that if DNA molecules are heated to 100°C, the two polynucleotide chains separate because the H bonds holding the base pairs of the opposite strands are disturbed. This process is known as ‘DNA denaturation’.
If the temperature is lowered, the complementary strands again unite. This is renaturation of DNA. These investigations also showed that isolated DNA strands could make hybrid or the mRNA. Several recent experiments indicate that only one strand of double stranded DNA molecule acts as template in mRNA synthesis.
Thus we can presume that mRNA is single stranded. mRNA shows base sequence that is complementary to that of template DNA. This process is called “transcription”.
This is just like replication. The only difference between the two processes is that in transcription thymine (T) is substituted by uracil (U) in mRNA, i.e., opposite adenine (A) of DNA comes U in mRNA. The mechanism of transcription is shown in Fig. 10.25. The enzyme RN A- polymerase has been found to catalyse synthesis of RNAs from ribonucleoside triphosphates (ATP, GTP, UTP, and CTP) on DNA template.
In eukaryotes, the mRNA migrates out of nucleus to the cytoplasm where the ribosomes are active in protein synthesis.
The main differences between prokaryotic mRNA and eukaryotic mRNA are as follows:
(i) mRNAs of prokaryotes are transcribed from several adjacent genes and as such the mRNAs are called polygenic or polycistronic mRNAs. In eukaryotes, however the mRNAs are transcribed on single genes hence they are called monogenic or monocistromic mRNA.
(ii) mRNAs in prokaryotes have short life times and are not processed whereas those of eukaryotes are processed and exhibit a range of life-times.
(iii) mRNAs of prokaryotes are transcribed on most of the genome where as those of eukaryotes are transcribed only on small fractions of genome.
At any one time less than 20% of the total cellular RNA is mRNA in either prokaryotic or eukaryotic cell.
Examination of mRNAs reveals that mRNA molecules are of variable length with fairly wide range of sedimentation coefficient (S values). That This results from the fact that the number of nucleotide pairs in a gene is related to the size of protein for which they code.
The general structure of nature mRNA molecule is shown in Fig. 10.33. All messenger RNAs are transcribed from the structural genes, i.e., genes that code for proteins.
Each mRNA has the following three main parts (Fig. 10.33):
(i) Leader sequence at 5′ end, the length of which may vary in different mRNAs and within this sequence is present the information for the ribosome to initiate protein synthesis at the correct position. The leader sequence is not involved in the translation process (protein synthesis).
(ii) Protein coding sequence following the leader sequence which is translated into the amino acid sequence of polypeptide, and
(iii) Trailer sequence at 3’end which is not translated and is variable in length in different mRNAs. The polycistronic mRNAs found in prokaryotes have leader and trailer sequence on the sides of the coding sequences.
The DNA of eukaryotic chromosome is organized into nucleosome structures. Many studies have shown that active chromatin is more sensitive to deoxyribonuclease attack than the non- transcribing chromatin. This indicates that transcribed segments of chromatin must have a different structural conformation than the inactive chromatin.
This is, however, not due to any major change in the nucleosome structure. Unlike, prokaryotic mRNAs, mRNAs, of eukaryotes are modified at both 5′ and 3′ ends. The modifications occur after transcription under the influence of specific enzymes.
Messenger RNA has the following structural features:
Cap:
At the 5′ end of the mRNA strand there occurs a cap which is blocked by methylated structure (any of the four nucleotides with 2-OH methyl ribose). Messenger RNA without cap shows poor binding of ribosomes and so the cap of mRNA influences the rate of protein synthesis (Fig. 10.34).
2. Non-coding region 1 (NCI):
Next to cap is non-coding region of 10-100 nucleotides which is rich in A and U bases and does not translate into protein.
3. The Initiation Codon:
This is AUG codon which initiates the polypeptide chain.
4. The Coding region:
Next to initiation codon comes the coding region which consists of about 1500 nucleotides on the average. This translates a particular protein molecule.
5. Terminating codon:
Next to the coding region there is termination codon which terminates protein synthesis on mRNA. In mRNA of eukaryotes, codons UAA, UAG and UGA act as termination codons.
6. Non-coding region (NC2):
This region lies next to termination codon which consists of 50-150 nucleotides. This region does not translate into protein. This region contains AAU sequence in all the cases.
7. Poly A sequence:
At the 3′ end there is a polyadenylate or poly-A (as AAAAA-) sequence which initially consists of200-250 nucleotides. Poly-A sequence is added in the nucleus before mRNA is transferred to cytoplasm. Poly-A sequence becomes reduced in extent with the age.
Split Genes and mRNA Production in Eukaryotes:
In eukaryotic nucleus a rapidly labelled RNA, called heterogenous nuclear RNA (hn RNA) species can be isolated which is different from rRNA and tRNA or their precursors. The hn RNA is heterodispersed in length and is much longer than the cytoplasmic wRNA and has both 5’and 3′ poly- A modifications characteristic of cytoplasmic mRNAs.
The hn RNA acts as pre- mRNA or precursor to the functional mRNAs, the former being processed to form latter in the nucleus.
A large fraction (approximately 90% of the hn RNA) is degraded and thus only a small portion of this is transported in the form of mRNA. In 1977, A.J. Jeffreys and R.A. Flavell discovered that some 600 base pairs (bp) sequence was inserted within the coding sequence for 146 aminoacids β globin chain in rabbit. The insert has been named an intervening sequence or intron and the coding sequences the exons.
The introns are transcribed and are not translated. An intron is a sequence in gene that separates coding sequences of genes. A large number of coding genes contain introns in eukaryotes. In higher eukaryotes, they are more extensive than in lower eukaryotes. Not all eukaryotic protein coding genes have introns. Histone genes of invertebrates are uninterrupted.
In general, there is no definite pattern to show as to which type of genes are interrupted and which type of genes are not. Simple model for the production of functional mRNA is presented in the Fig. 10.35. At first, both coding sequences and introns are transcribed but each intron is subsequently removed in such a way that the adjacent coding genes are spliced together to form a continuous sequence.
The process is summarized as under:
(i) RNA polymerase binds to the promoter and transcribes on the DNA continuously until a termination is reached, i.e., leader trailer and coding sequences as well as introns are transcribed.
(ii) Transcript becomes capped at the 5′ end. The transcript is then cleaved near the 3′ end and a poly-A tail is added to the new 3′ end to produce a precursor mRNA (pre-mRNA) which is longer than the mature mRNA because of presence of introns. The pre-mRNA molecule constitutes hn RNA
(iii) The pre-mRNA molecule is processed to remove introns which are looped out and sub squinty the loops are removed by nucleases. The new adjacent coding sequences are then linked or spliced together. This is called splicing of introns. It is known that all introns start with GU and end with AG and these sequences are presumably crucial for the correct splicing.
Informosome:
Many investigators like Spirin, Beltisina and Lerman (1965), Perry and Kelley (1968) and Henshaw (1968) have reported that in certain eukaryotic cells the messenger RNA does not enter the cytoplasm as a naked RNA strand but often remains ensheathed by certain proteins.
Spirin has proposed a term informosome for mRNA-protein complex. The proteins of informosome provide stability to the mRNA and protect mRNA from degrading action of ribonuclease enzyme.