ADVERTISEMENTS:
This article throws light upon the three types of cloning vectors used in recombinant DNA technology.
The three types of cloning vectors are: (1) Bacteriophage (2) Phagemids and (3) Cosmids.
Type # 1. Bacteriophage as Cloning Vectors:
The plasmid based vectors used for cloning DNA molecules generally carry up to 10 kb of inserted DNA. However, for the formation of library, it is often helpful to be able to maintain larger pieces of DNA. For this reason, E. coli virus (Bacteriophage, phage) lambda (λ) has been developed as a cloning vehicle. In its life cycle, bacteriophage λ infects E. coli and after injection of the viral DNA, two possibilities exist.
ADVERTISEMENTS:
Bacteriophage λ can enter a lytic cycle, which after 20 minutes lead to the lysis of host cells and the release of about 100 phage particles. Alternatively, the injected bacteriophage λ DNA can be integrated into the E. coli chromosome (DNA) as a prophage and can be maintained more or less indefinitely (Lysogeny stage).
However, under conditions of nutritional or environmental stress, the integrated bacteriophage λ DNA can be excised and enter a lytic cycle. The bacteriophage λ DNA is about 50 kb in length, of which approximately 20 kb is essential for the integration excision (I/E) events. For forming genomic libraries, 20 kb of DNA can be replaced with 20kb of cloned DNA.
Cloning Vectors based on the Bacteriophage Lambda (λ):
Derivatives of the genome of bacterioplage lambda have been constructed to serve as cloning vectors. Transfection or transduction is used to introduce such vectors into E. coli.
Two properties of the lambda genome make it suitable for use as a vector:
ADVERTISEMENTS:
1. Only about 50% of the 50 genes of lambda are essential for its replication and for lysis of the host cell. Most of these non essential genes are located together in a cluster around the middle of the genome.
2. Lambda genome is packaged inside the phage head by what is known as the ‘head-full mechanism’. This means that not only there is an upper limit of the amount of DNA that is packaged inside the phage head, but there is a lower limit also. Effective packaging takes place only when a minimum amount of DNA is present, i.e., 35 kb (kilo = thousand base).
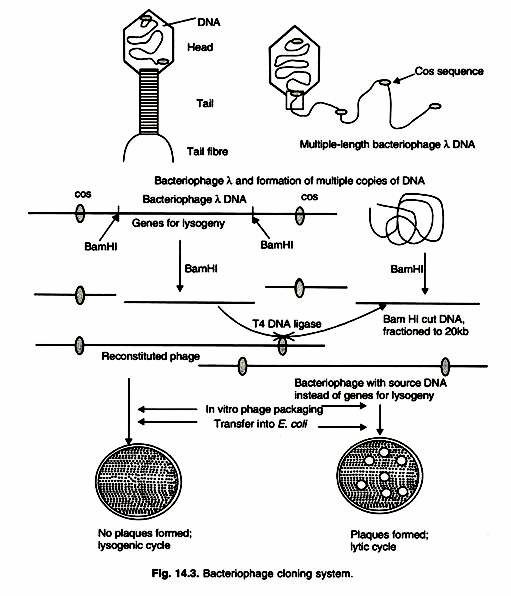
An infective bacteriophage λ consists of a tubular protein tail with a few tail fibres and a protein head. The production and assembly of heads and tails, and packaging of DNA are highly coordinated sequence of events. The DNA within head of a λ phage is a 50 kb linear molecule with a 12-base, single stranded extension at the 5′ end. These extensions are called cohesive (cos) ends, because they contain sequences that are complementary to each other.
In E. coli, these cos ends base pair to form a circular DNA. DNA replication from the circular DNA creates a linear form of λ DNA that is composed of several contiguous lengths of 50 kb units. Each new assembly is filled with 50 kb DNA (Fig. 14.3).
Inserting Type Lambda (λ) Vectors:
A lambda cloning vector can therefore be constructed by deletion (in vivo or in vitro by restriction deletion) of a part of the non-essential region such that the remainder is not less than 35kb. Other mutations are also introduced such that restriction sites in the enemies regions are eliminated. A segment of foreign DNA can be cloned in an unique restriction site in the non-essential region, the only condition being that the vector and the insert together would not to be more than 53 kb long. Such vectors are termed “insertion vectors”.
Some of the Charon vectors are examples of this type of vectors. Bacteriophage X cloning vector has two Bam HI sites that flank the I/E region. When this DNA is cut with Bam HI, three segments are produced. The middle segment I/E region, which is replaced by cloned DNA of 20 kb size. The source is cut with Bam HI, and DNA pieces that are 15 to 20 kb in length are isolated. The two DNA samples (phage and source) are combined and incubated with T4 DNA ligase.
Then empty bacteriophage heads and tail parts are added. Under these conditions 50 kb unit of DNA are packed in to the heads, and infective phages are produced. Other products from ligation reaction cannot be packed, because they are either too large (>52 kb) or too small (>38 kb). Recombinant bacteriophage λ can undergo lytic cycle only in an E. coli strain that does not allow reconstituted phage λ (non-recombinant) with intact I/E regions to grow.
Recombinant phage is maintained by lytic cycles in fresh E. coli cultures. Bacteriophage libraries can be screened by using either DNA probes or immunological assays. For this purpose, individual lytic zones are tested (compared to bacterial colonies in plasmid cloning vactors).
Substitution Type Lambda (λ) Vector:
ADVERTISEMENTS:
The second type of lambda vector is of the substitution type, the example being the lambda gt vectors and the EMBL vectors. These vectors have two Eco R1 sites or two BamHI sites in the non-essential region.
On digestion with Eco Rl, at least three piece(s) are produced, two terminal ones containing the essential regions and the central piece(s) containing the non-essential genes.
The central piece (s) is separated out by sucrose density gradient sedimentation and replacing by the foreign segment to be cloned. The limits of the size of foreign DNA that can be cloned in the lambda gt vector is 1-14 kb and in the Charon 4 vector is 8.2-22.2 kb.
Such a replacement cloning vector has an advantage over the insertion vectors. The terminal pieces by themselves if joined by DNA ligase, do not make up 35 kb and hence cannot be packaged. Packaging occurs only when a segment of foreign DNA gets cloned between the two terminal pieces of the vector and hence no separate selection for recombination molecules to is necessary.
Type # 2. Phagemids as Cloning Vectors:
A phagemids is a hybrid of a plasmid and a filamentous coliphage that can be propagated in either form. The coliphage could be either of the three virtually identical phages, M13 fd or f1. These are male specific phages that contain single stranded circular DNA as their genome. Upon infection of E. coli by the bacteriophage, double stranded DNA is first formed as the replicative intermediate.
ADVERTISEMENTS:
Finally single stranded DNA is packaged into the virion. Both the replication origins of the plasmid and the coliphage are incorporated in the phagemid. The auxiliary replication functions necessary in trans for the coliphage replication are not, however, incorporate in the phagemid. Hence, replication from the coliphage origin can take place only in the presence of a helper phage.
Otherwise, replication takes place from the plasmid replication origin. The pBLUESCRIPT phagemids have both Col E, (pMB 9 like) origin and the filamentous phage f1 origin. The cloning is done in any of the multiple cloning sites in the double stranded circular DNA of the plasmid form of the vector. This is introduced in to E.coli by transformation and the synthesis of single stranded DNA from the phage fl origin is induced by superinfection with a helper phage.
The single stranded DNA formed is packaged in to the phage rods because of the presence of the phage packaging signals as well in the phagemid. Direct base sequencing can be undertaken using the single stranded DNA isolated from the virions secreted from the E. coli cells.
ADVERTISEMENTS:
Depending on the orientation of the f1 replication origin in the phagemid, either the (+) strand or the (-) strand of the phagemid is replicated in presence of the helper phage. The pEMBL phagemids are similar to the pBLUESCRIPT phagemids.
Phagemids that have replication origin of pUC plasmids and of the M13 bacteriophage have also been developed and are available under the trade name of LITMUS vectors. Replication in the circular double stranded plasmid form uses the pUC replication origin.
Cloning is done in the multiple cloning sites located in the lacZ gene and the usual blue/white selection is available. Here also replication of the single stranded phage DNA form is induced by super infecting with the M13 helper phage.
Type # 3. Cosmids as Cloning Vectors:
Plasmid vectors are not suitable for cloning DNA fragments very much larger than their own size, as the transformation frequency fall beyond acceptable limits and cloned fragments or their parts very often get deleted. Takagi and co-workers observed as early as 1976 that the presence of the cohesive end site cos λ from the bacteriophage lambda DNA in a plasmid allows it to be packaged in vivo into virus particles. The interesting finding was that the in vivo packaging mechanism would be select DNA molecules of the full size of the lambda genome (~48.5 kb).
ADVERTISEMENTS:
Making use of this finding, cosmid vectors were first developed in 1978 by J. Collins and coworkers to facilitate cloning of larger DNA fragments in plasmids. Extracts of lambda lysogens have been successfully used for in vitro packaging of the lambda capsids. An example of a commonly used cosmid is pHV79 which is nothing but pBR322 containing the cohesive end site cos λ and which can accommodate up to 45 kb sized inserts.
A great advantage of such a cosmid vector is that:
(1) Gene libraries consisting of a smaller number of clone members can span the whole genome of an organism. For example, the genome of Escherichia coli can be accommodated in just 120 cosmids.
(2) Other advantages are that large gene can be studied intact and genetic linkage studies can be carried out at the molecular level.
(3) An important practical advantage of a cosmid is that background molecules which do not have the intact and genetics linkage studies can be carried out at the molecular level.
(4) An important practical advantage of a cosmid is that background molecules which do not have inserts or have smaller inserts are eliminated during packaging. This is not possible to achieve with plasmid cloning vectors.
ADVERTISEMENTS:
(5) Besides, the frequency of transformation of the lambda capsids with an in vitro packaging extract is much higher than the transformation frequency of plasmids.
Cosmid cloning vectors can carry 40 kb of cloned DNA and can be maintained as plasmids in E. coli. Cosmids combine the properties of plasmids and bacteriophage λ vectors. The commonly used cosmid pLFR-5 (6kb size) has two cos sites (cos ends) from bacteriophage λ separated by a Sea I restriction endonuclease site, a multiple cloning sequence with six unique sites (Hind III, PstI, Sail, BamHI, SmaI, and EcoRI), an origin of DNA replication (ori) and a tetracycline resistance (Jet) gene. This cosmid carry about 40 kb of cloned DNA.
For this vector, pieces of DNA that are approximately 40 kb in length are purified by sucrose density gradient configuration from a partial digestion of source DNA with Bam HI. The pFLR-5 DNA is cleaved first with Seal and then with Bam HI. The two DNA samples are mixed and ligated. Some of the ligand products will have a 40kb DNA piece inserted between the two fragments that are derived from the digestion of the pLFR-5 DNA.
The molecules formed by joining will be about 50 kb long, with cos sequences that are about 50 kb apart. Therefore, these DNA constructs can be successfully packaged into bacteriophage λ heads in vitro (as described above, phage λ head accommodate only 50kb DNA). After formation of complete phage, the DNA is delivered by infection into E. coli.
During phage packaging, cos ends are cleaved. Once inside the bacteria, the cos ends base pair and form a circular DNA molecules (Fig. 14.4). This circular form is stable, so the cloned DNA can be maintained as a plasmid-insert DNA construct because the vector contains a complete set of plasmid functions. Moreover, the tetracycline resistance gene allows colonies that carry the cosmid to grow in presence of tetracycline; non-transformed cells are sensitive to tetracycline and die.
The following are the steps for construction of a cosmid library:
ADVERTISEMENTS:
(i) Cleavage of the genome by partial digestion with restriction endonuclease,
(ii) Sizing of the fragments by gel electrophoresis or velocity centrifugation;
(iii) Cleavage of the cosmid vector and treatment with phosphate to minimize polycomid formation;
(iv) Ligation of the genomic DNA and the cosmid DNA;
(v) Packaging the ligated DNA into infectious phage particles;
(vi) Transduction into E. coli.
Therefore, it is evident that different vectors have different capacity of carry foreign DNA (Table 14.2).